Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
An alternative metric for measurement of methlyation levels are M-values. β-values can be easily converted to M-values using the following equation:
M-value = log2( β / (1 - β))
An M-value close to 0 for a CpG site indicates a similar intensity between the methylated and unmethylated probes, which means the CpG site is about half-methylated. Positive M-values mean that more molecules are methylated than unmethylated, while negative M-values mean that more molecules are unmethylated than methylated. As discussed by Du and colleagues, the β-value has a more intuitive biological interpretation, but the M-value is more statistically valid for the differential analysis of methylation levels.
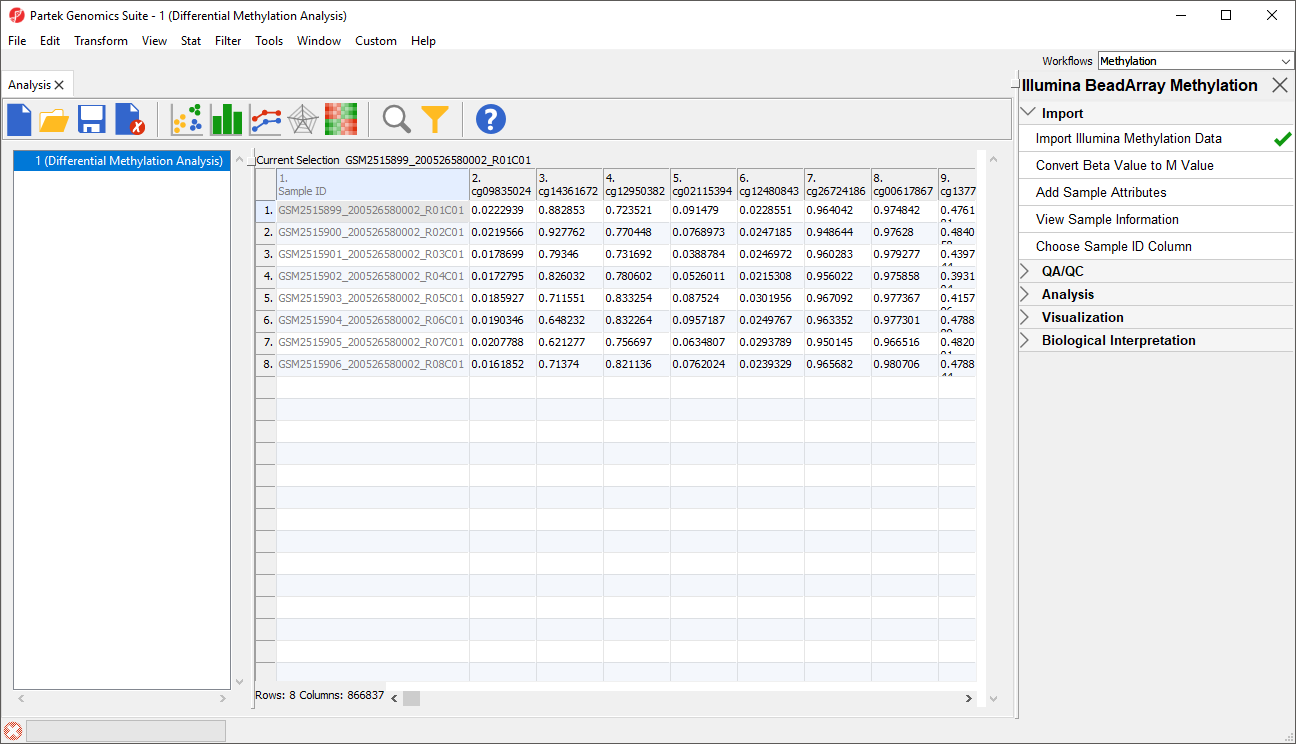
Because we are performing differential methylation analysis, we need to convert our data to from β-values to M-values.
- Select Convert Beta Value to M Value from the Import section of the Illumina BeadArray Methylation workflow
The original data (β-values) will be overwritten.
- Select () from the icon bar to save the current spreadsheet
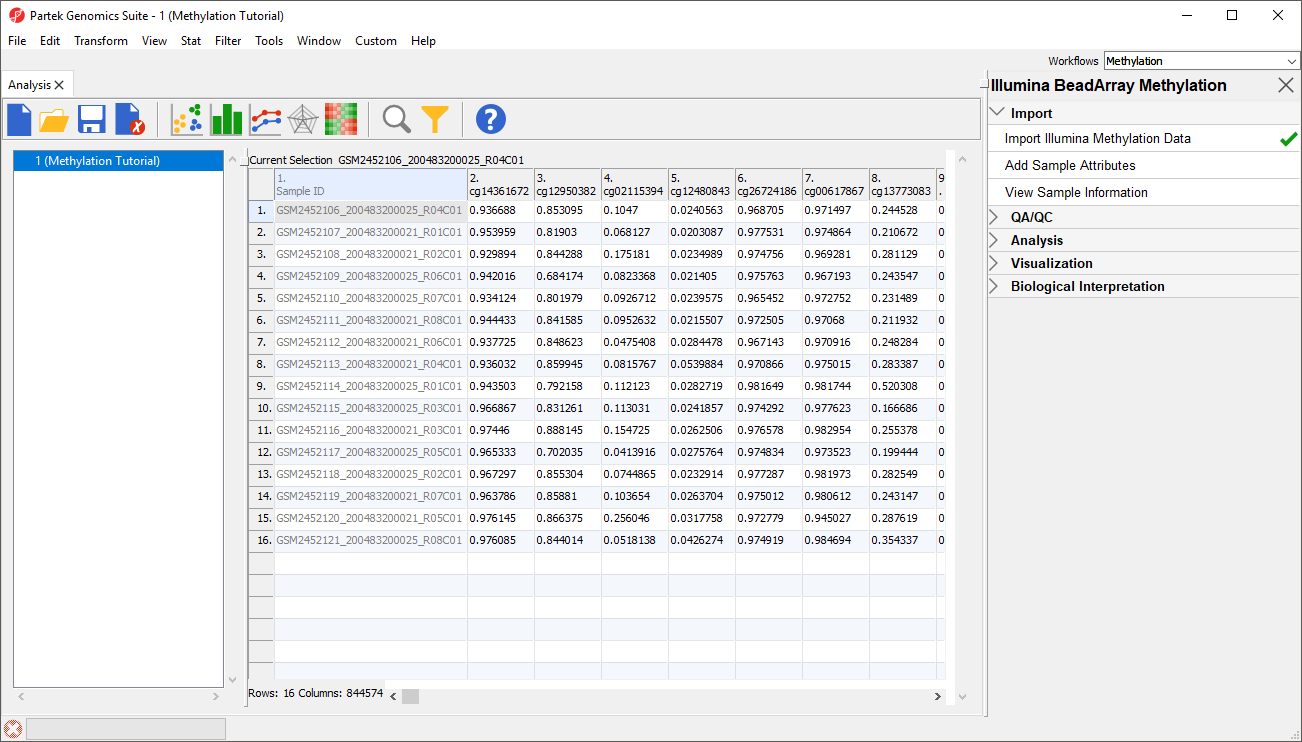
Before we can perform any analysis, the study samples need to be organized into their experimental groups.
...
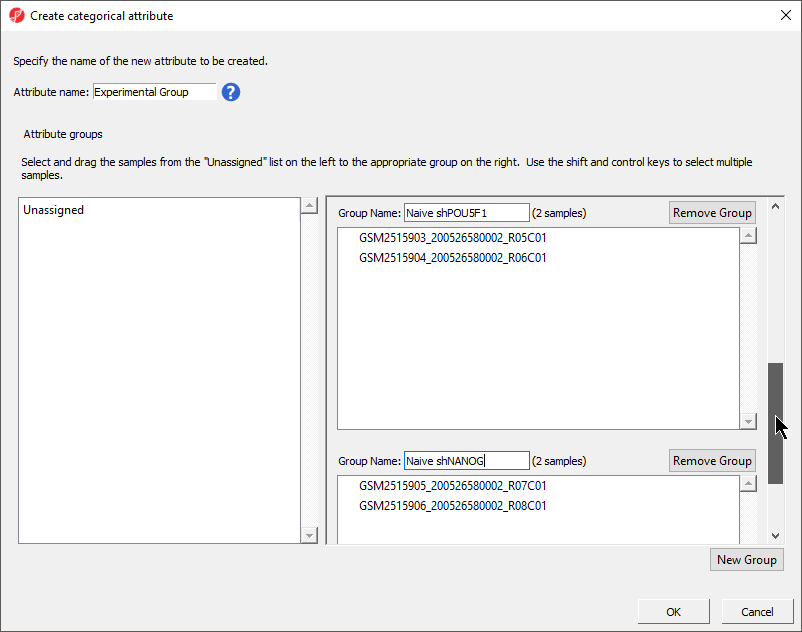
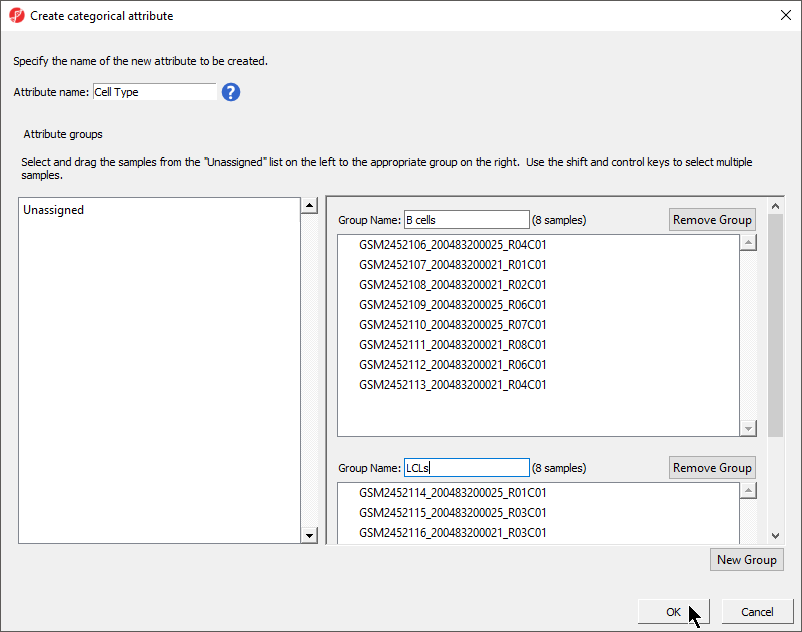
The Create categorical attribute dialog allows us to create groups for a categorical attribute. By default, two groups are created, but additional groups can be added.
- Set Attribute name: to StateCell Type
- Rename the groups Primed and Naivegroups B cells and LCLs
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Group NameCell Type | |
|---|---|---|
| GSM2515899GSM2452106_200526580002_200483200025_R04C01 | B cells | |
| GSM2452107_200483200021_R01C01 | PrimedB cells | |
| GSM2515900GSM2452108_200526580002200483200021_R02C01 | PrimedB cells | |
| GSM2515901GSM2452109_200526580002200483200025_R03C01R06C01 | NaiveB cells | |
| GSM2515902GSM2452110_200526580002_R04C01 | Naive | |
| GSM2515903_200526580002_R05C01 | Naive | |
| GSM2515904_200526580002_R06C01 | Naive | |
| GSM2515905_200526580002_R07C01 | Naive | |
| GSM2515906_200526580002_R08C01 | Naive200483200025_R07C01 | B cells |
| GSM2452111_200483200021_R08C01 | B cells | |
| GSM2452112_200483200021_R06C01 | B cells | |
| GSM2452113_200483200021_R04C01 | B cells | |
| GSM2452114_200483200025_R01C01 | LCLs | |
| GSM2452115_200483200025_R03C01 | LCLs | |
| GSM2452116_200483200021_R03C01 | LCLs | |
| GSM2452117_200483200025_R05C01 | LCLs | |
| GSM2452118_200483200025_R02C01 | LCLs | |
| GSM2452119_200483200021_R07C01 | LCLs | |
| GSM2452120_200483200021_R05C01 | LCLs | |
| GSM2452121_200483200025_R08C01 | LCLs |
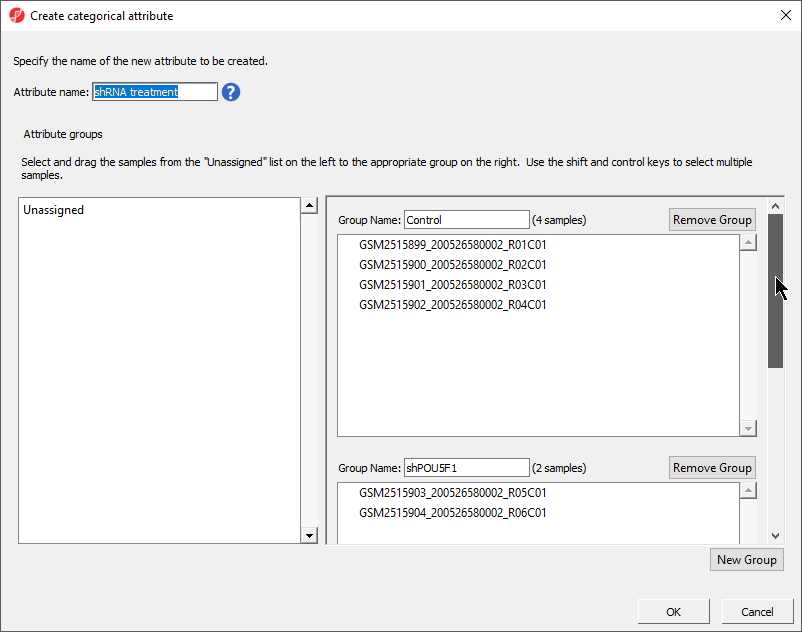
There should now be two groups with eight samples in each group (Figure 43).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select OK
- Select Yes from the Add another categorical attribute dialog
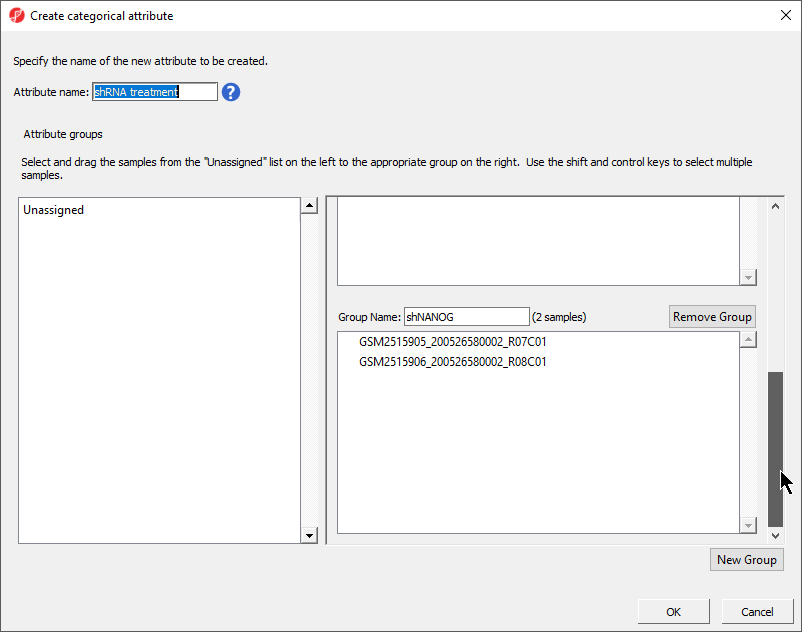
- Set Set Attribute name: to shRNA treatmentSelect New Group to add an additional groupto Gender
- Rename the three groups Control, shPOU5F1, and shNANOGgroups Male and Female
- Drag and drop the samples from the Unassigned list to their groups as listed in the table below
| Sample ID | Group NameGender | ||
|---|---|---|---|
| GSM2452106_ | 200526580002_200483200025_R04C01 | Female | |
| GSM2452107_200483200021_R01C01 | ControlFemale | ||
| GSM2452108_ | 200526580002200483200021_R02C01 | ControlMale | GSM2515901|
| GSM2452109_ | 200526580002200483200025_ | R03C01R06C01 | ControlFemale |
| GSM2515902GSM2452110_200526580002_R04C01 | Control | ||
GSM2515903_200526580002_R05C01 | shPOU5F1 | ||
GSM2515904_200526580002_R06C01 | shPOU5F1 | ||
GSM2515905_200526580002_R07C01 | shNANOG | ||
GSM2515906_200526580002_R08C01 | shNANOG200483200025_R07C01 | Female | |
| GSM2452111_200483200021_R08C01 | Female | ||
| GSM2452112_200483200021_R06C01 | Female | ||
| GSM2452113_200483200021_R04C01 | Male | ||
| GSM2452114_200483200025_R01C01 | Female | ||
| GSM2452115_200483200025_R03C01 | Female | ||
| GSM2452116_200483200021_R03C01 | Male | ||
| GSM2452117_200483200025_R05C01 | Female | ||
| GSM2452118_200483200025_R02C01 | Female | ||
| GSM2452119_200483200021_R07C01 | Female | ||
| GSM2452120_200483200021_R05C01 | Female | ||
| GSM2452121_200483200025_R08C01 | Male |
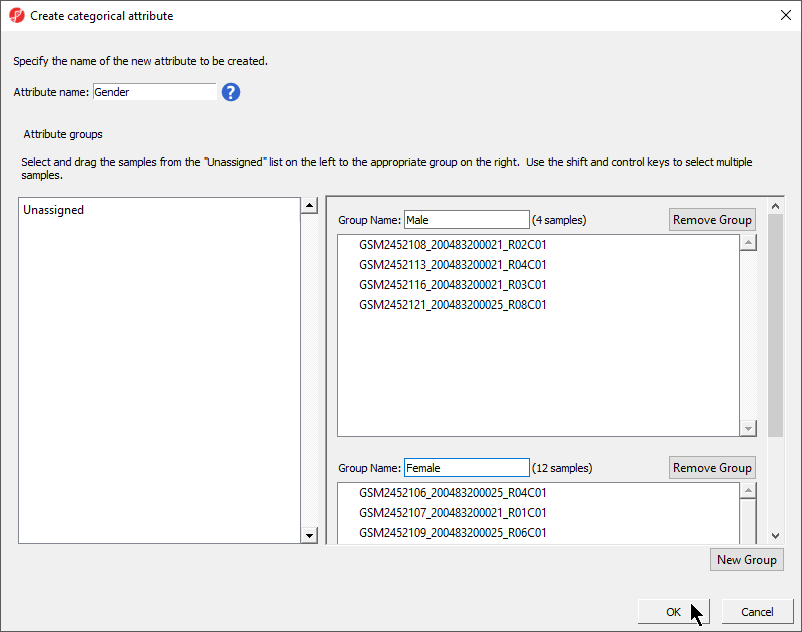
There should now be three two groups with four samples in each group Male and twelve samples in Female (Figure 54).
| Numbered figure captions | ||||||
|---|---|---|---|---|---|---|
|
| |||||
- Select OK
- Select Select No from the Add another categorical attribute dialog
- Select Yes to save the spreadsheet
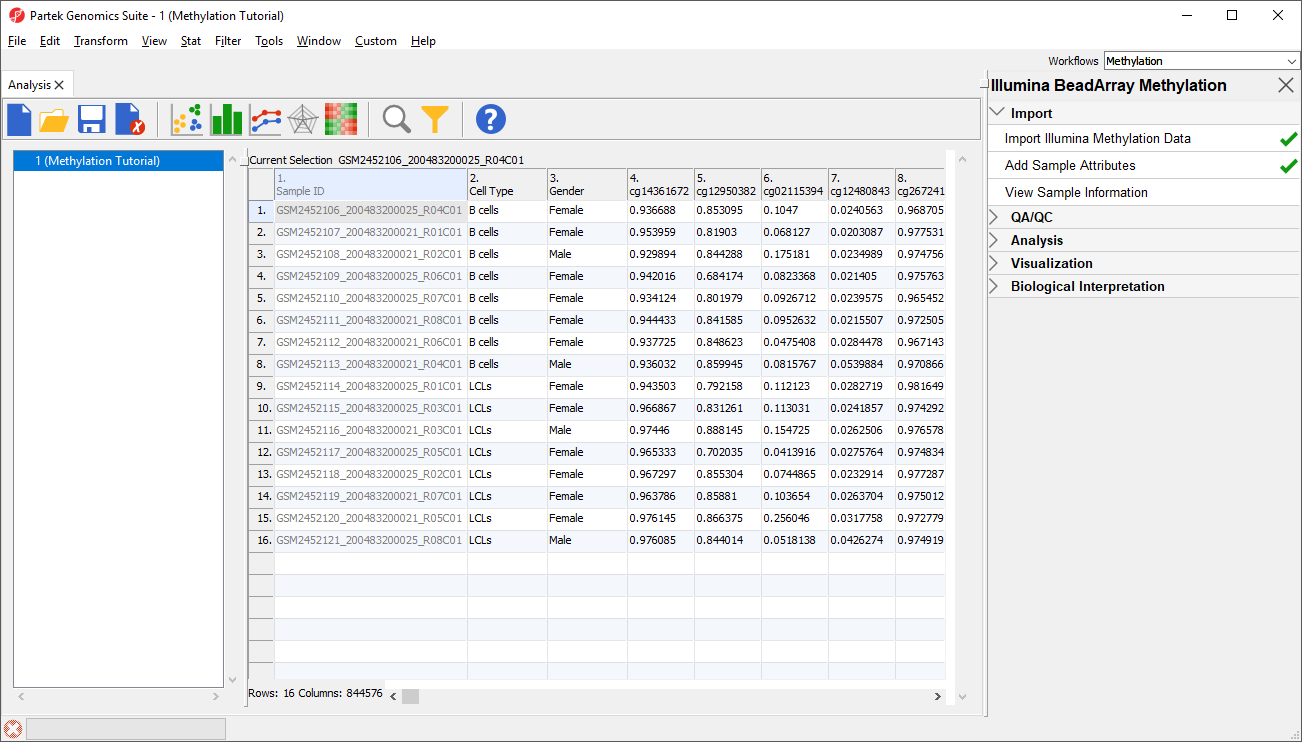
Two new column as columns have been added to spreadsheet 1 (Differential Methylation Analysis) with the state cell type and shRNA treatment gender of each sample (Figure 65).
| Numbered figure captions | |
|---|---|
|
...
|
...
| |
| Page Turner | ||
|---|---|---|
|
...