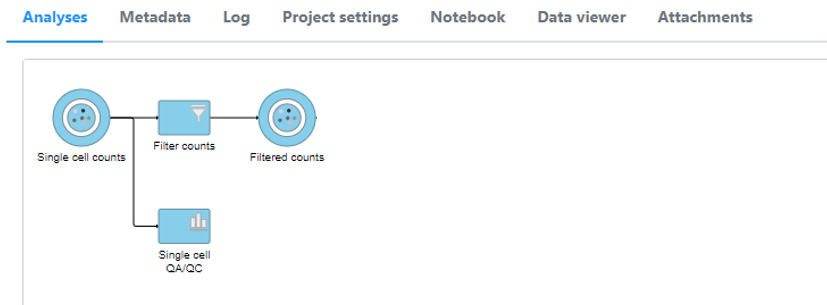
Page History
...
- Click a Single cell counts data node
- Click the QA/QC section of the task menu
- Click Single cell QA/QC
By default, all samples are used to perform QA/QC. You can choose to split the sample and perform QA/QC separately for each sample.
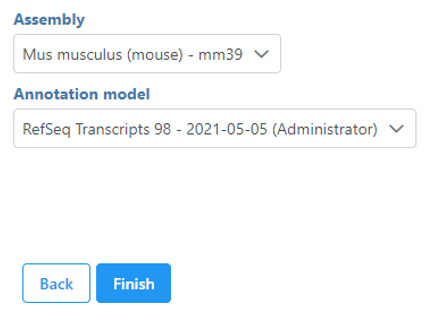
If your Single cell counts data node has been annotated with a gene/transcript annotation, the task will run without a task configuration dialog. However, if you imported a single cell counts matrix without specifying a gene/transcript annotation file, you will be prompted to choose the genome assembly and annotation file by the Single cell QA/QC configuration dialog (Figure 1). Note, it is still possible to run the task without specifying an annotation file. If you choose not to specify an annotation file, the detection of mitochondrial counts will not be possible.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
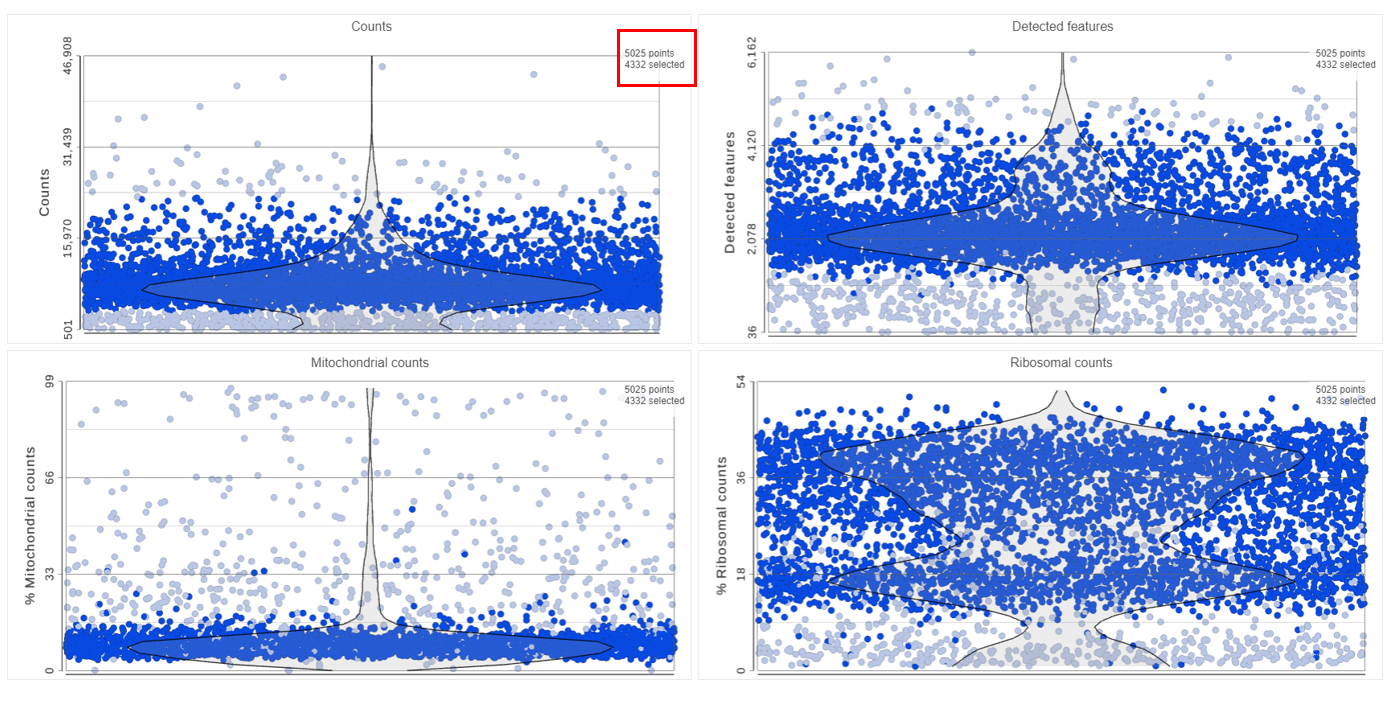
The Single cell QA/QC task report opens in a new data viewer session. Four dot and violin plots showing the value of every cell in the project are displayed on the canvas: counts per cell, detected features per cell, the percentage of mitochondrial counts per cell, and the percentage of ribosomal counts per cell (Figure 2).
...
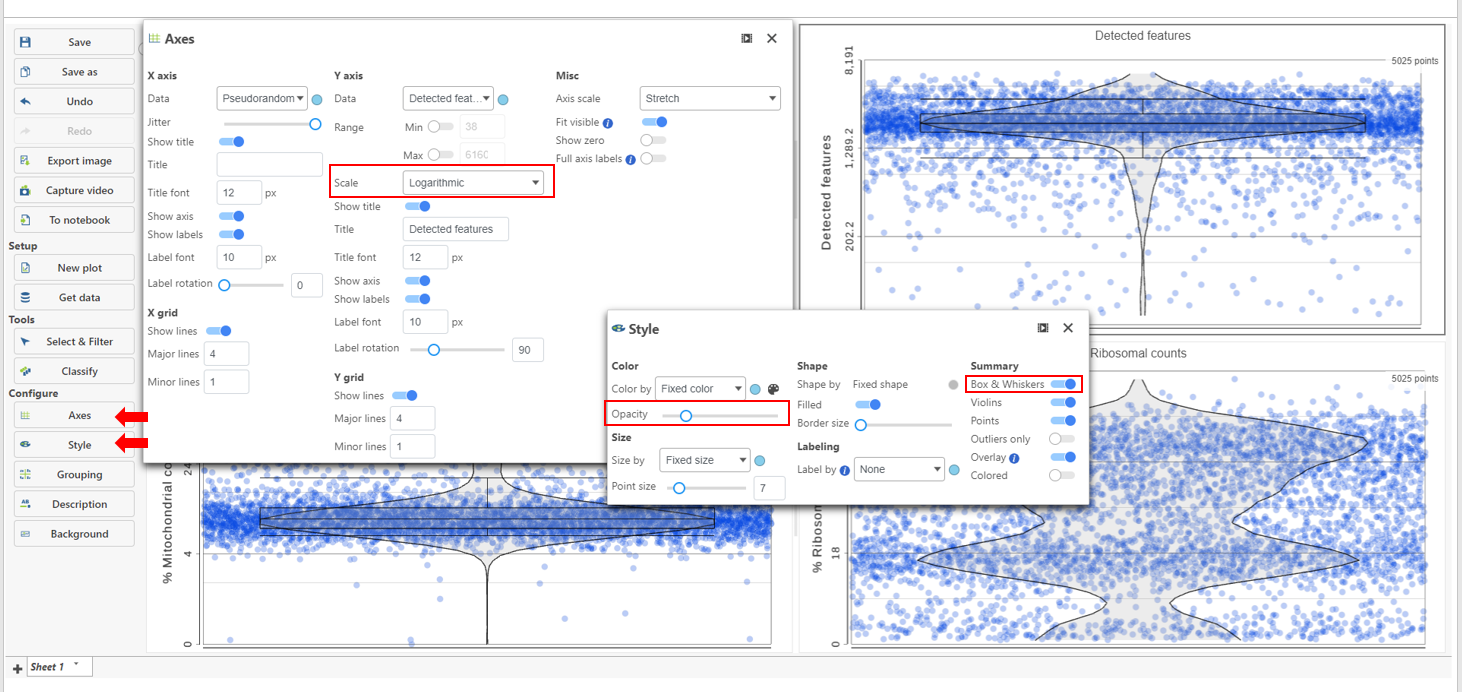
The appearance of a plot can be configured by selecting a plot and adjusting the Configure settings in the Configuration cards panel on the left (Figure 4). Here are some suggestions, but feel free to explore the other options available:
- Open the Color card and reduce the Opacity using the slider. For data sets with very many cells, it may be helpful to decrease the dot opacity to better visualize the plot density.Open the Scale card and Axes and change the Y-axis scale to Logarithmic. This can be helpful to view the range of values better, although it is usually better to keep the Ribosomal counts plot in linear scale.
- Open the Summary card and switch on Style and reduce the Color Opacity using the slider. For data sets with very many cells, it may be helpful to decrease the dot opacity to better visualize the plot density.
- Within Style switch on Summary Box & Whiskers. Inspecting the median, Q1, Q3, upper 90%, and lower 10% quantiles of the distributions can be helpful in deciding appropriate thresholds.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
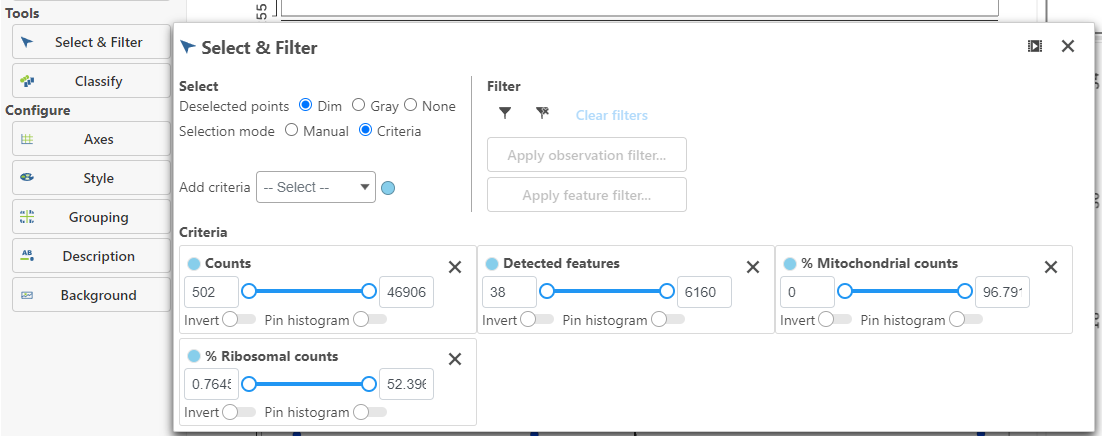
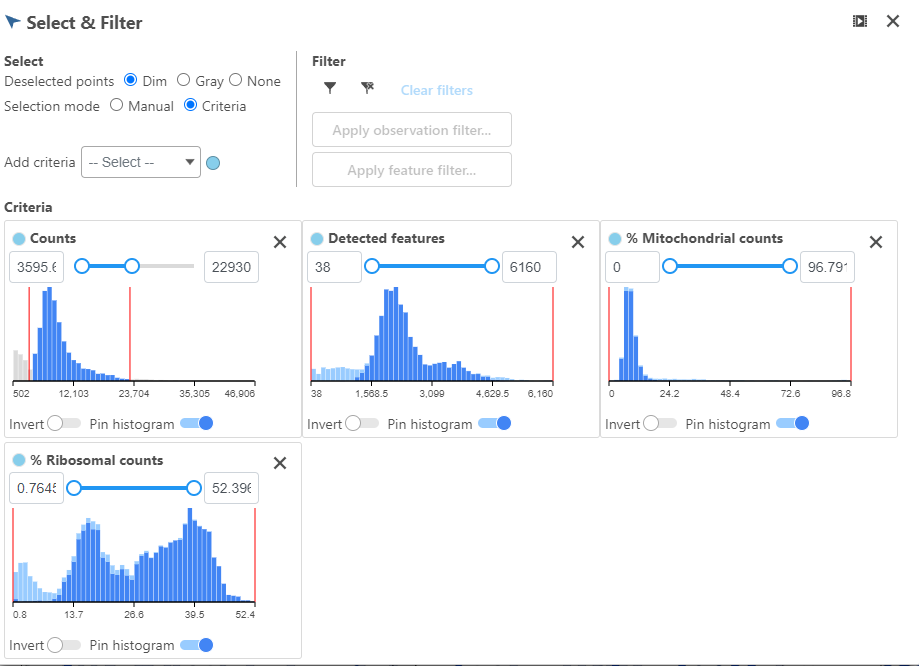
High-quality cells can be selected using the Selection card on the right Select & Filter, which is pre-loaded with three the selection rulescriteria, one for each quality metric (Figure 5).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
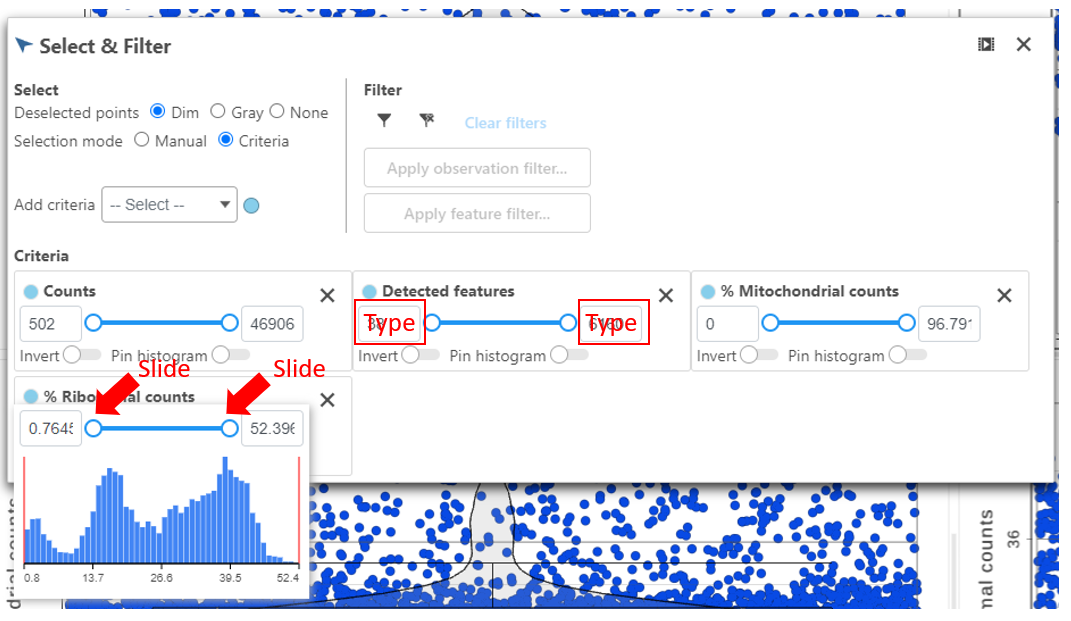
Hovering the mouse over one of the selection rules criteria reveals a histogram showing you the frequency distribution of the respective quality metric. The minimum and maximum thresholds can be adjusted by clicking and dragging the sliders sliders or by typing directly into the text boxes for each selection criteria (Figure 6).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Alternatively, Pin histogram to view all of the thresholds can be set by typing directly into the text boxes in each selection rule distributions at one time to determine thresholds with ease (Figure 7).
| Numbered figure captions | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| |||||||
Adjusting the selection criteria will select and deselect cells in all three plots simultaneously. Depending on your settings, the deselected points will either be dimmed or gray. The filters are additive. Combining multiple filters will include the intersection of the three filters. The number of cells selected is shown in the figure legend of each plot (Figure 8).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To filter the high-quality cells, click the the include selected cells icon in Filter in the Filtering card in the top right of Select & Filter, and click Apply observation filter... (Figure 9).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
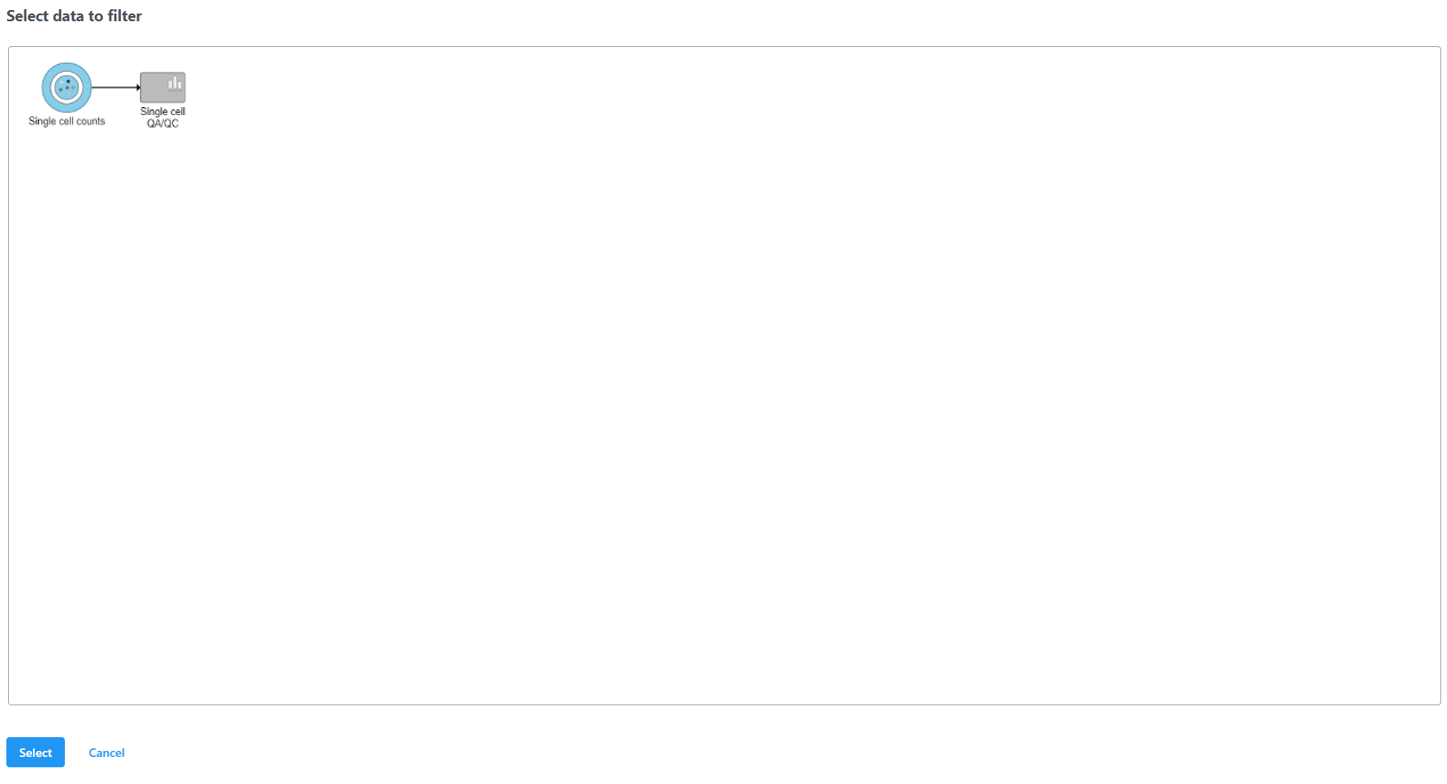
Select the input data node for the filtering task and click Select (Figure 10).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new data node, Filtered counts, will be generated under the Analyses tab (Figure 11).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
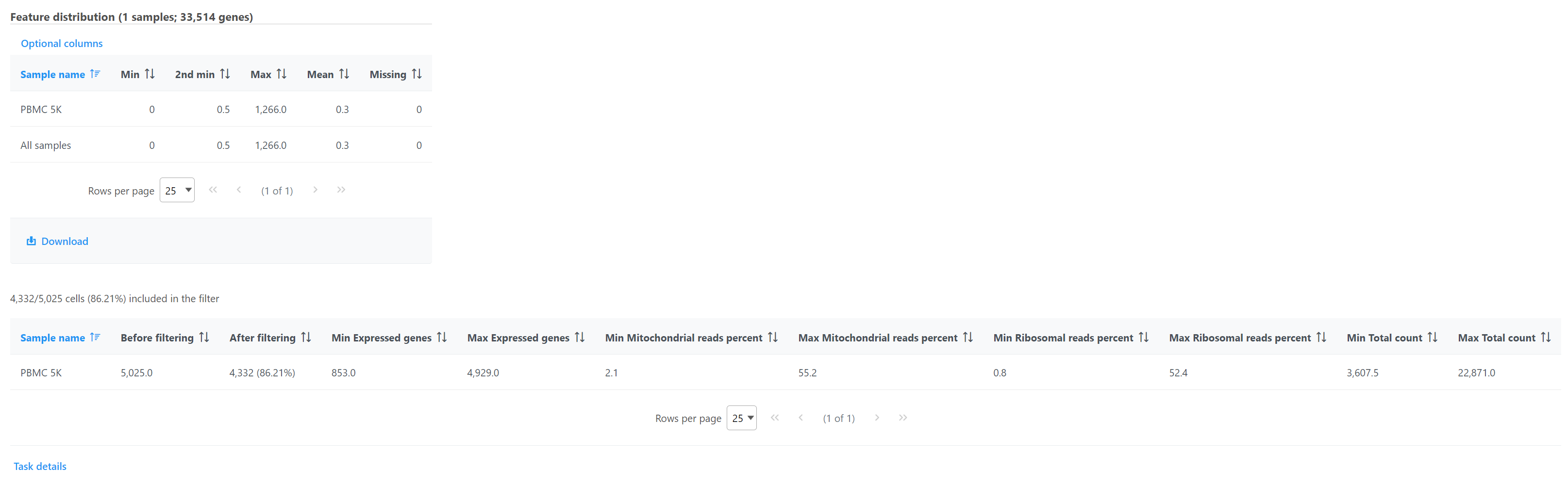
Double click the Filtered counts data node to view the task report. The report includes a summary of the count distribution across all features for each sample; a detailed breakdown of the number of cells included in the filter for each sample; and the minimum and maximum values for each quality metric (expressed genes, total counts, etc) across the included cells for each sample (Figure 12).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
...