Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
What is Trim tags?
The Trim tags task allows you to process sequences unaligned read data with adaptors, barcodes, and UMIs using a Prep kit file that specifies the structure of the adapters, barcodes, UMIs, and insert sequences configuration of these elements in your NGS reads.
Running Trim tags
- Click an Unaligned reads data node
- Click the Pre-alignment QA/QC section of the toolbox
- Click Trim tags
There are two three parameters to configure - Prep kit and , Keep untrimmed, and Map feature barcodes.
Selecting Keep untrimmed will generate a separate unaligned reads data node with any reads that do not match the structure specified by the prep kit. This option is off by default, to save on disk space. Selecting Map feature barcodes is only necessary for processing protein data from 10x Genomics' Feature Barcoding assay (v3+ chemistry). For single cell gene expression data, leave this option unchecked.
Partek distributes prep kits for processing several types of data - Drop-seq, :
- 10x Chromium Single Cell 3' v2
...
- 10x Chromium Single Cell 3' v3
- 10x Chromium Single Cell 5'
- Drop-seq
- Lexogen QuantSeq FWD-UMI
- Bio-Rad SureCell WTA 3'
- Fluidigm C1 mRNA Seq HT IFC
...
- Rubicon Genomics ThruPLEX Tag-seq
...
- 1CellBio inDrop
If your data is from one of these sources, you can select the appropriate option in the Prep kit drop-down menu. If the data is from another source, you can build a custom prep kit file to process your data.
- Choose a Prep kit from the drop-down menu
Click Finish to run Trim tags (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The output of Trim tags is a Trimmed reads data node (and an . An additional Untrimmed reads data node will be generated if the Keep untrimmed untrimmed option was selected).
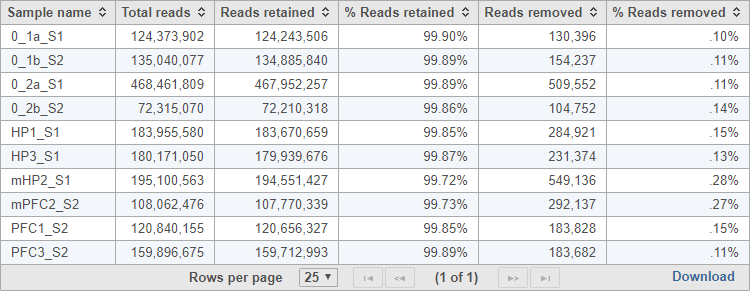
The task report provides a table with the total reads, reads retained, % reads retained, reads removed, and % reads removed for each sample (Figure 2). You can click Download at the bottom of the table to save a text file copy to your computer.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Building a custom prep kit
...
The Prep kit builder interface will load (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
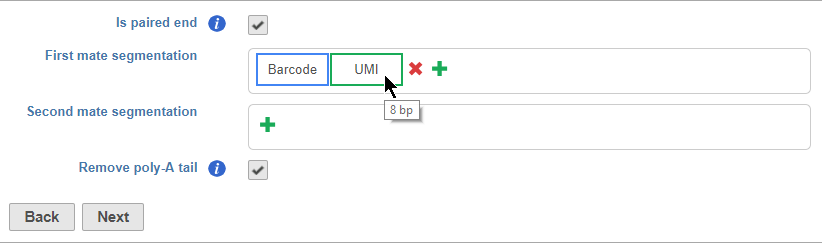
Is paired end - select to switch from single end to paired end FASTQ files (Figure 5). If you choose paired end, the First mate will correspond to the _R1 FASTQ file and the Second mate will correspond to the _R2 FASTQ file.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
To use a file, choose File for Sequences and then click Choose File (Figure 6). Use the file browser to choose a FASTA file from your local computer.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
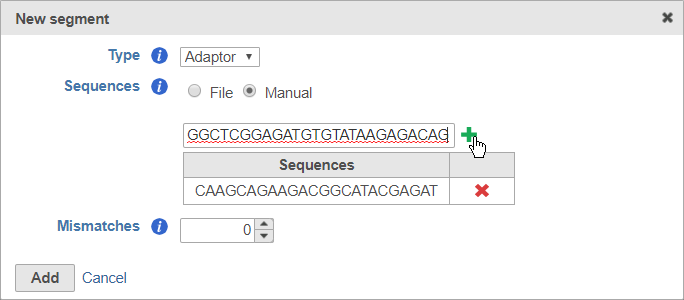
To enter the sequences manually, choose Manual for Sequences then type or paste the adaptor sequences into the text field and click to add the adaptor (Figure 7). You must click for the adaptor sequence to be included. You can remove any adaptor you have added by clicking .
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Including a UMI in your prep kit will allow you to access a downstream task that uses UMI information , Deduplicate UMIsfor removing PCR duplicates. For more information about the Deduplicate UMIs task, please see our UMI Deduplication in Partek Flow white paper. While Note that while the UMI sequence will be trimmed, a record of the UMI sequence for each read is retained for use by this downstream taskstask.
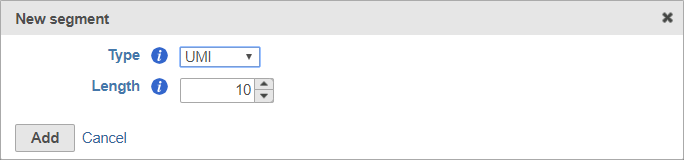
When adding a UMI segment to your prep kit, you can specify the length of your UMIs (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Adding a barcode segment to a prep kit allows you to access downstream tasks that use barcode information, including Filter including Filter barcodes and Quantify barcodes to annotation model (Partek E/M). While the barcode sequence will be trimmed, a record of the barcode sequence for each read is retained for use by downstream tasks.
Like adaptors, barcodes can be specified using a file or manually specified, but you can also choose to designate any a segment of arbitrary length in the sequence as the barcode. This is useful if you do not have a specific set of known barcodes.
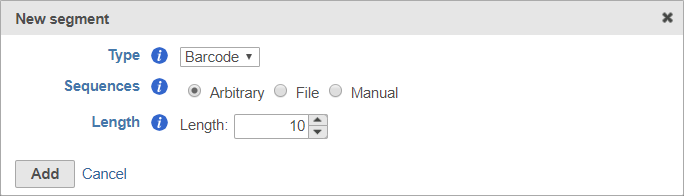
To set the barcode to an arbitrary segment of fixed length, choose Arbitrary and specify the barcode length (Figure 9).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
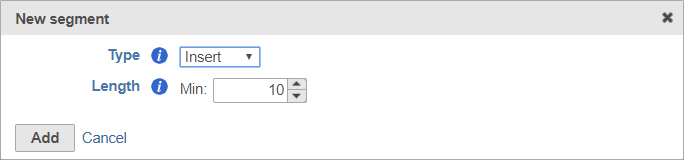
The insert is the sequence retained after trimming in the Trimmed reads data node. For example, in RNA-Seq, this would be the mRNA sequence. Every prep kit must include an insert segment. You can specify the minimum size of the insert section using the Length field (Figure 10). Reads shorter than the minimum length will be discarded.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Remember to click Add to add the new segment to your prep kit.
...
Ordering segments
Segments are placed from 35' to 53' in the read in the order they are added. You should add the 35' segment first and add additional elements in order of their position in the read. Segments will appear in the Segmentation sections as they are added. You can mouse over a segment to view its details (Figure 11).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Custom prep kit example
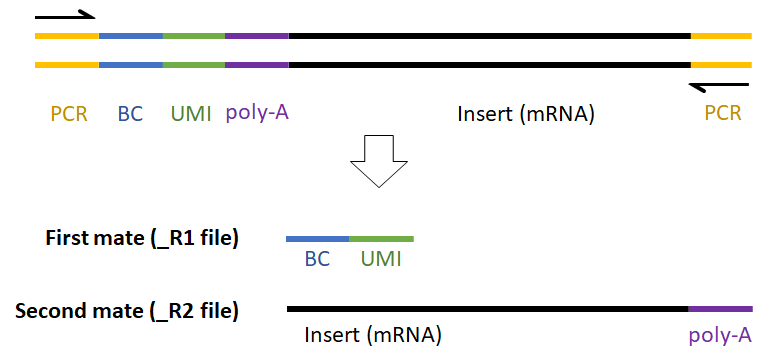
For example, the expected read structure (Figure 12) and a completed prep kit for a standard Drop-seq library prep are shown below (Figure 13).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
- Click Next to complete your prep kit
Managing prep kits
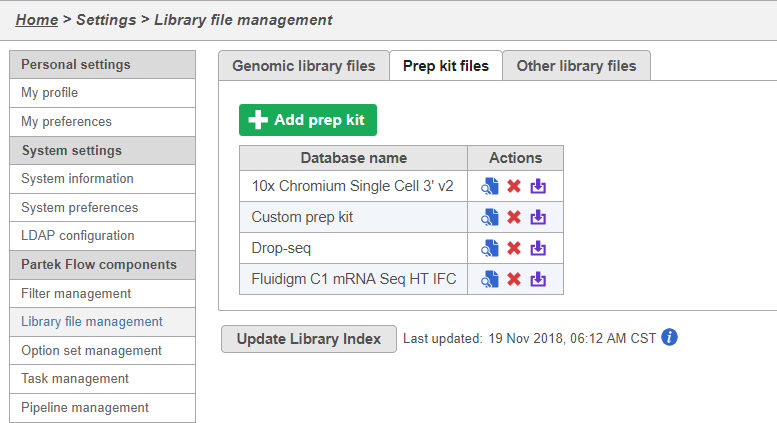
You can manage saved prep kits by going to Home > Settings > Library file management and opening the Prep kit files tab (Figure 14).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Prep kits download as a .zip file. This Prep kit .zip file can be imported into Partek Flow by selecting Import from a file when adding a new prep kit. Select the .zip file when importing, do not unzip the file.
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...