Page History
...
Binding sites for the DNA-binding protein of interest are indicated by peaks of enriched sequencing read density. Because each single-end read only covers one end of an immunoprecipitated DNA fragment, enriched regions will have two adjacent peaks of increased sequencing read density from reads on the forward and reverse strands. To merge these peaks, each read is extended in the 3' direction by the effective fragment length, converting reads into estimated fragments. Overlapping estimated fragments are then merged into peaks. For peak detection, the genome is divided into bins of a user-defined size and the number of estimated fragments that fall in each bin is calculated. A zero-truncated negative binomial model, appropriate for data is fitted to the bin counts and all regions that are enriched above a user-defined false discovery rate (FDR) are called as peaks.How are peaks calculated from reads in Partek Genomics Suite?
Using the effective fragment length calculated by Cross Strand-Correlation, each read is extended in the 3' direction by the effective fragment length and overlapping extended reads are merged into a single peakpeaks. For paired-end reads, the distance between paired reads is used as the fragment length and overlapping fragments are merged into peaks. For peak detection, Partek Genomics Suite divides the genome is divided into windows of a user-defined size and counts the number of fragments whose mid-points fall within each window is counted. A statistical test is then applied model for expected read density (a zero-truncated negative binomial) is used to determine which peaks are significant. See significantly enriched over a user-defined false discovery rate (FDR). See the ChIP-Seq white paper for for more information on the peak-finding algorithm and tips for setting the Fragment extension and window sizes.
...
The Peak Detection dialog will open. We will configure Configure the dialog as shown (Figure 1).
...
- Select Maximum average fragment size for Fragment Extensions
- Set Maximum average fragment size to 111
Maximum Your choice for Maximum average fragment size is based on your experimental design: the size of the fragment pulled-down by immunoprecipitation, the fragment sizes produced by DNA fragmentation, the fragment length selected by size exclusion, or the effective fragment length calculated by Cross Strand-Correlation. If you have used an antibody that binds DNA as the control antibody such as an IgG control, you could use different fragment lengths for each sample based on its effective fragment length by selecting the Individual maximum fragment sizes option. Here, we have chosen the effective fragment length of 111 base pairs calculated using Cross Strand-Correlation.
- Select Reference sample from Reference sample
- Select mock from the Reference sample drop-down menu
- Set Set the window size to (base pairs) to 111
The peak detection algorithm divides the genome into windows to find windows with enriched for reads based on an FDR cut-off value. Here, we have chosen to match the window and individual maximum fragment sizes.
...
The Peak Cut-off FDR determines the cut-off for calling peaks. Setting a lower value demands greater differences between mock and chip samples fora for a peak to be called; a false discovery rate of 0.001 anticipates 1 false positive per 1000 peaks called.
...
Optimal peak detection settings are dependent on your experimental design and data so fine tuning may be required. Because transcription factor binding sites tend to have localized and sharp clusters of reads, the window size used during the analysis of a transcription factor study can be left relatively small, approximately the same as the average fragment length, and the option to allow for gaps between enriched windows does not need to used. Additionally, Region in the window with most reads could also be selected to report a more narrow region for each peak call. Conversely, histone modification peaks tend to be subtle and diffuse. To analyze histone modification ChIP-seq data, larger window sizes, combining neighboring windows into larger windows using Within using Within a gap distance of, and reporting entire regions using Entire region, spanning all merged windows might be appropriate.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
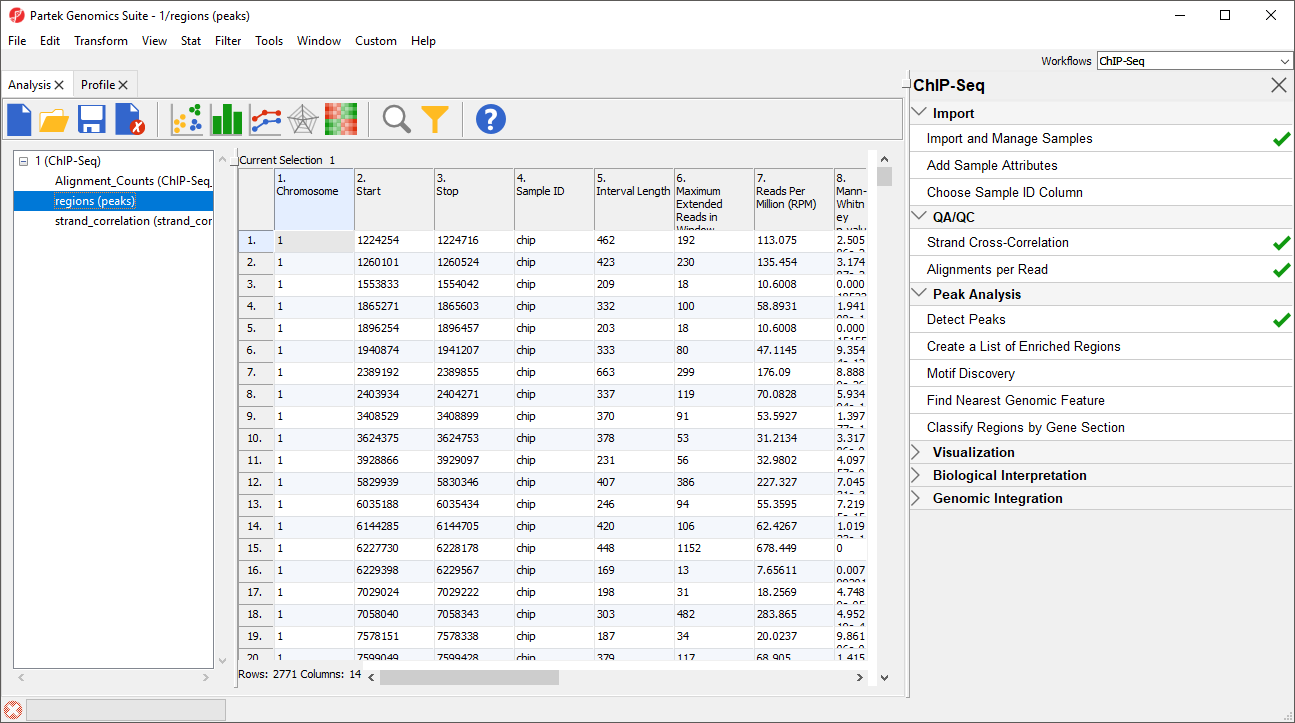
A few of the columns contents merit clarification.
...
This spreadsheet is sorted by chromosome number and genomic location. Each row represents one genomic region of peak enrichment. The columns are:
Column 1. Chromosome gives the chromosome location of region
Column 2. Start gives the start of region (inclusive)
Column 3. Stop gives the end of region (exclusive)
Column 4. Sample ID gives the sample containing the enriched region
Column 5. Interval length gives the length of the region, Start - Stop, in base pairs
Column 6. Maximum Extended Reads in Window gives the greatest number of extended reads in any of the windows of a region
Column 7. Reads per Million (RPM) divides the total number of aligned reads in the sample (in millions). This helps you compare peaks across samples, especially when there is a large difference in the number of aligned reads between samples.
Column 8. Mann-Whitney p-value identifies the separation between forward and reverse peaks for single-end reads using the Mann-Whitney U-test. Lower p-values indicate better separation. This p-value can be used if there was no control sample or to eliminate regions called due to PCR bias.
Columns 9-10. Total reads in region gives the total number of non-extended reads for each sample in the given genomic region. One column for each sample.
Column 11. p-value(Sample ID. vs. mock) compares the sample specified in column 4 to the reference sample for this genomic region using a one-tailed binomial test. A low p-value means there are significantly more reads in the sample specified in column 4 than in the mock sample. This column is only included if a reference sample is specified.
Column 12. scaled fold change (Sample ID vs. mock) compares intensity of signal between the sample specified in column 4 to the reference sample in the given genomic region. The fold-change is scaled by a ratio of the number of reads for each sample (IP vs. control) on a per-chromosome basis. Scaled fold changes >1 indicate more enrichment in the IP-sample than in the control sample. This column is only included if a reference sample is specified.
Columns 13 -14. <Sample ID> overlap percent gives the fraction of the given genomic region that overlaps a called peak region from the indicated sample. For example, the values of 100% in column 13 and 0% in column 14 indicate regions detected in the chip sample, but not in the mock sample. Similarly, regions with the value of 100% in column 14 were detected in the mock sample.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|