Page History
...
- Click a Taxonomic data node
- Choose Alpha & beta diversity from the Metagenomic section of the toolbox
- (optional) If you wish to compare the alpha diversity values between groups of samples, choose the factors to set up the ANOVA. This can e be helpful if you want to see if there is a significant difference in species diversity between predefined groups. This is optional, so you can skip this if you want.
- Click Finish
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
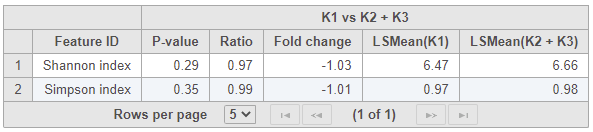
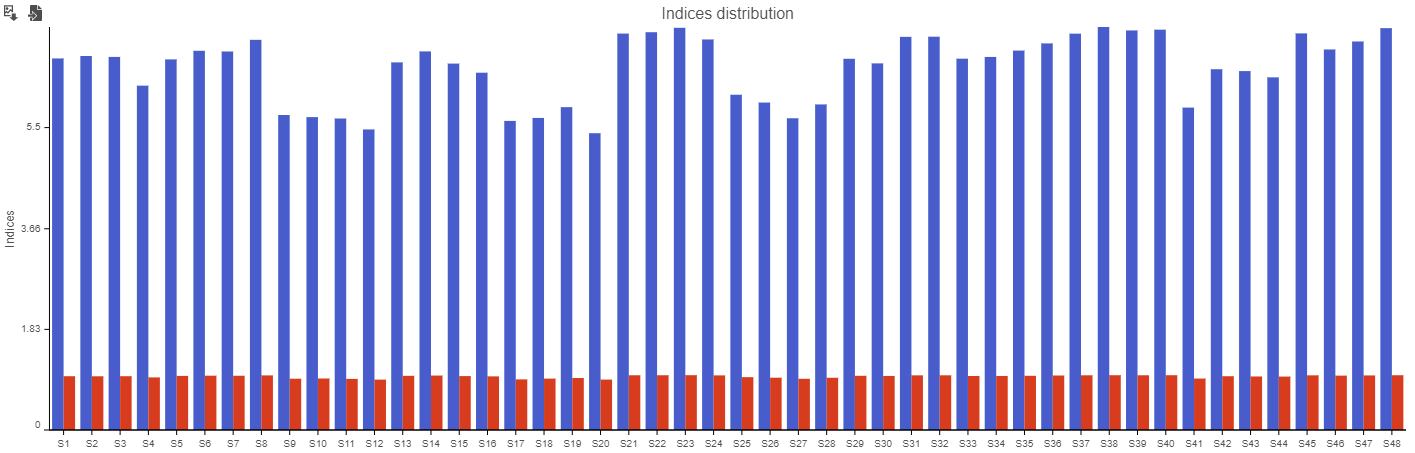
At the bottom, there is a bar chart summarizing the Shannon and Simpson metrics for each sample (Figure 34).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
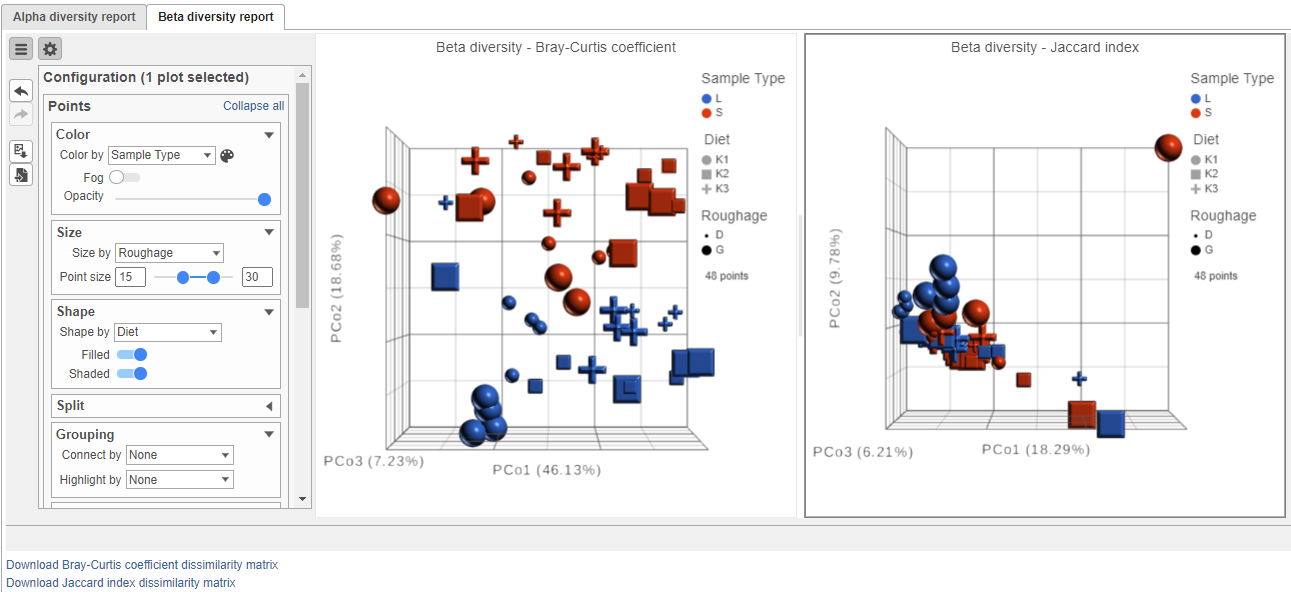
The beta diversity results are presented in a Data viewer session, with two PCoA plots on the canvas. One plot shows the clustering based on the Bray-Curtis Coefficient coefficient distance metric. The other shows the clustering on the Jaccard index distance metric. In both plots, each point is a different sample and they cluster together based on how similar their overall metagenomic profiles are. Points very close together are similar, points far apart are different. The Configuration panel on the left can be used to customize the plots.
To obtain the table of pairwise comparisons (dissimilarity matrix) for each distance metric, click the appropriate hyperlink below the Data viewer. The table will be downloaded as a tab-delimited text file. For both distance metrics, the values range from 0 to 1. The higher the value, the more different that pair of samples are. The lower the value, the more similar they are.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To obtain the table of pairwise comparisons (dissimilarity matrix) for each distance metric, click the
References
- Shannon CE: A mathematical theory of communication. Bell System Technical Journal 1948, 27.
- Simpson EH: Measurement of diversity. Nature 1949, 163: 688.
- Bray JR, Curtis JT: An ordination of upland forest communities of southern Wisconsin. Ecol . Monogr. 1957, 27.
- Beals E: Bray-Curtis ordination: an effective strategy for analysis of multivariate ecological data. Adv . Ecol . Res.Res 1984, 14.
- Jaccard P: Lois de distribution florale. Bulletin de la Socíeté Vaudoise des Sciences Naturelles 1902, 38.
- Jaccard P: The distribution of the flora in the alpine zone. New Phytologist 1912, 11:2
- Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R: Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2010, 7:10
...