Page History
Seurat v3[1] introduced new methods for the integration of multiple single-cell datasets, no matter whether they were collected from different individuals, experimental conditions, technologies, etc. Seurat 3 integration method aims to first identify 'anchors' between pairs of datasets. These 'anchors' are then used to harmonize the datasets. Its standard workflow is provided in Partek Flow.use a subset of the data as reference for the integrate analysis. The method integrates all other data with the reference subset. The subset can be one sample or a subgroup of samples defined by the factor attribute.
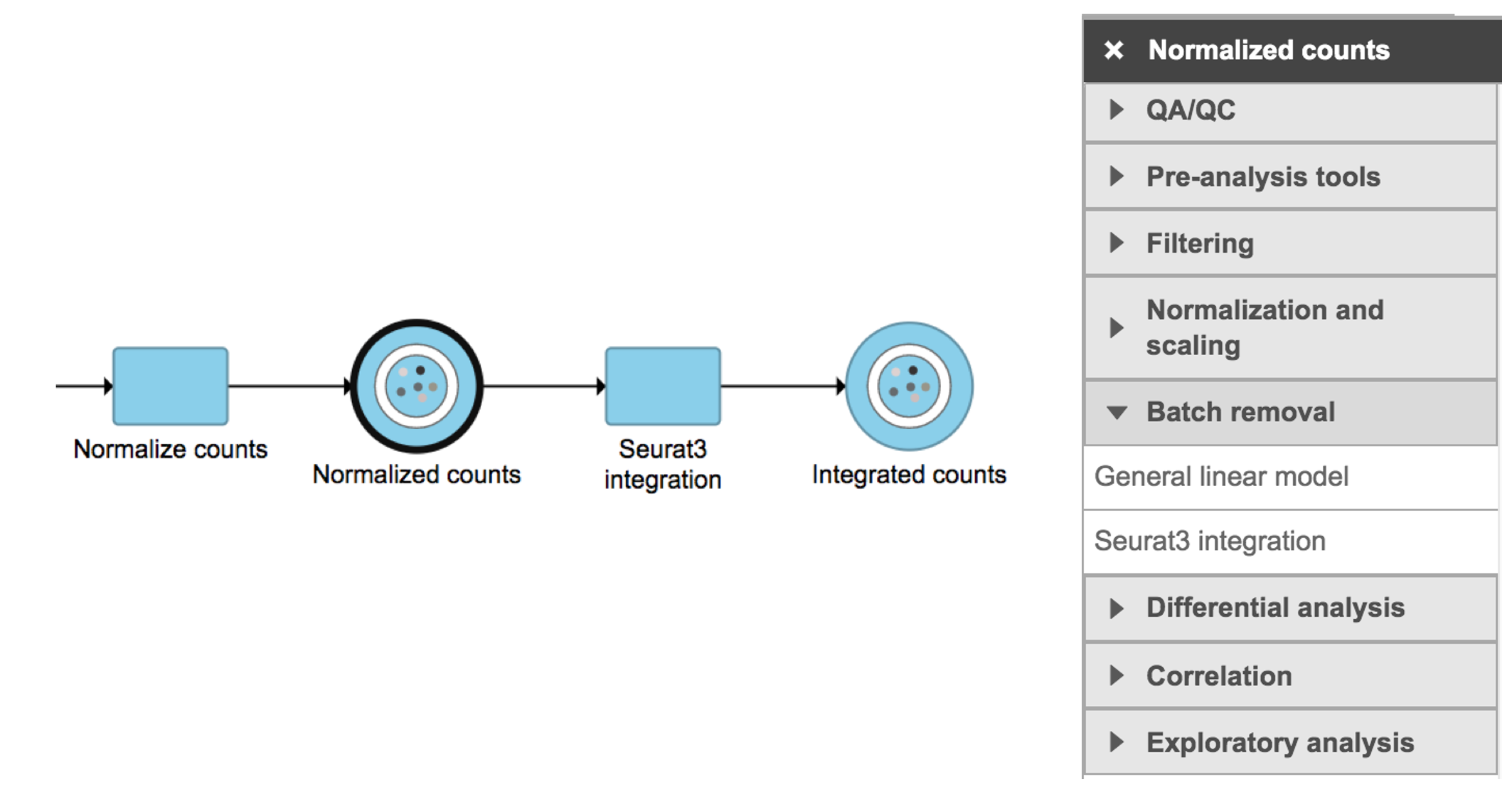
Seurat3 integration in Flow can be invoked in Batch removal section if a Normalized counts data node is selected (Figure 1).
Figure 1. Seurat3 integration task in Batch removal section in Flow.
...
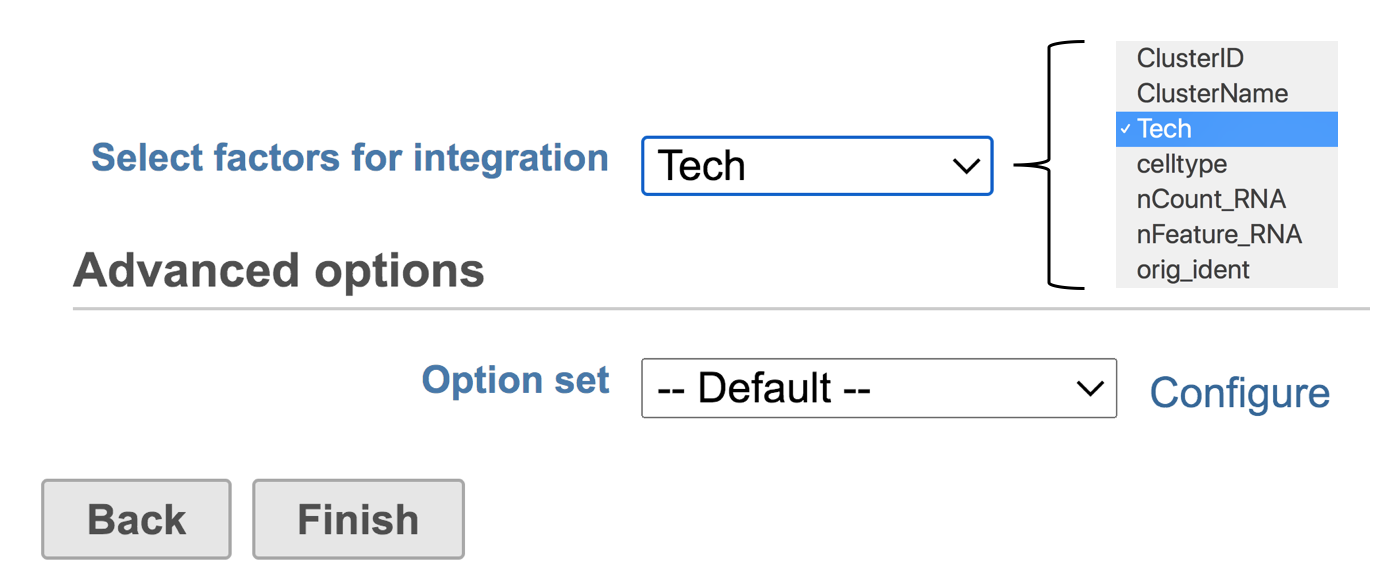
- Select Tech from the dropdown list
- Click Finish
Figure 2. Interface of Seurat3 integration in Flow. Example attributes are indicated in the drop-down list.
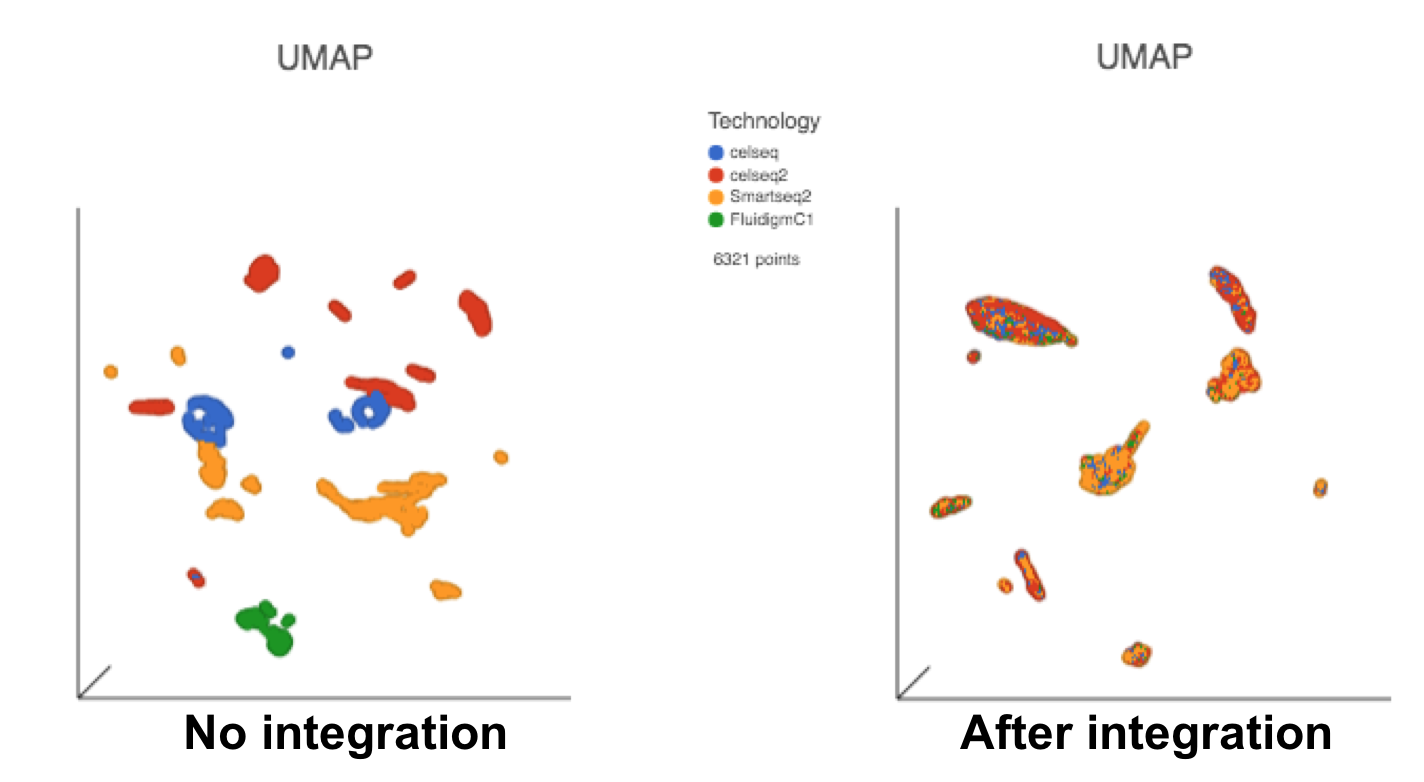
The output of Seurat3 integration is a new data node - Integrated counts (Figure 1). We can then use this new integrated matrix for downstream analysis and visualization (Figure 3).
Figure 3. UMAP displays the cells colored by Tech attribute before(left) and after(right) Seurat3 integration. Both images in the figure share the same legend.
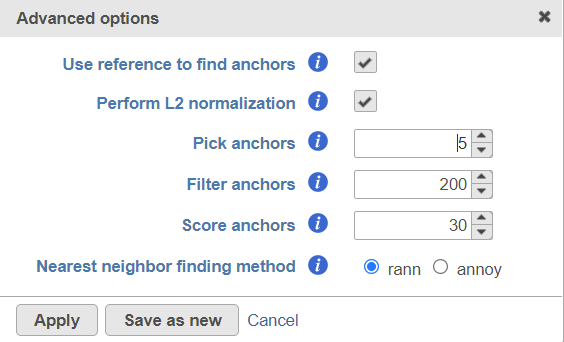
Users can click Configure to change the default settings In Advanced options (Figure 4).
Figure 4. Advanced configure options for Seurat3 integration in Flow.
Use reference to find anchors: when this box is checked, the first group of the selected attribute is used as reference to find anchors. To use a different group as reference, change the order of subgroups of the attribute in the attribute management page on Data tab. When the box is unchecked, anchors will be identified by comparing all pairs of subgroups, this option is very computationally intensive.
Perform L2 normalization: Perform L2 normalization on the CCA cell embeddings after dimensional reduction.
...