Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
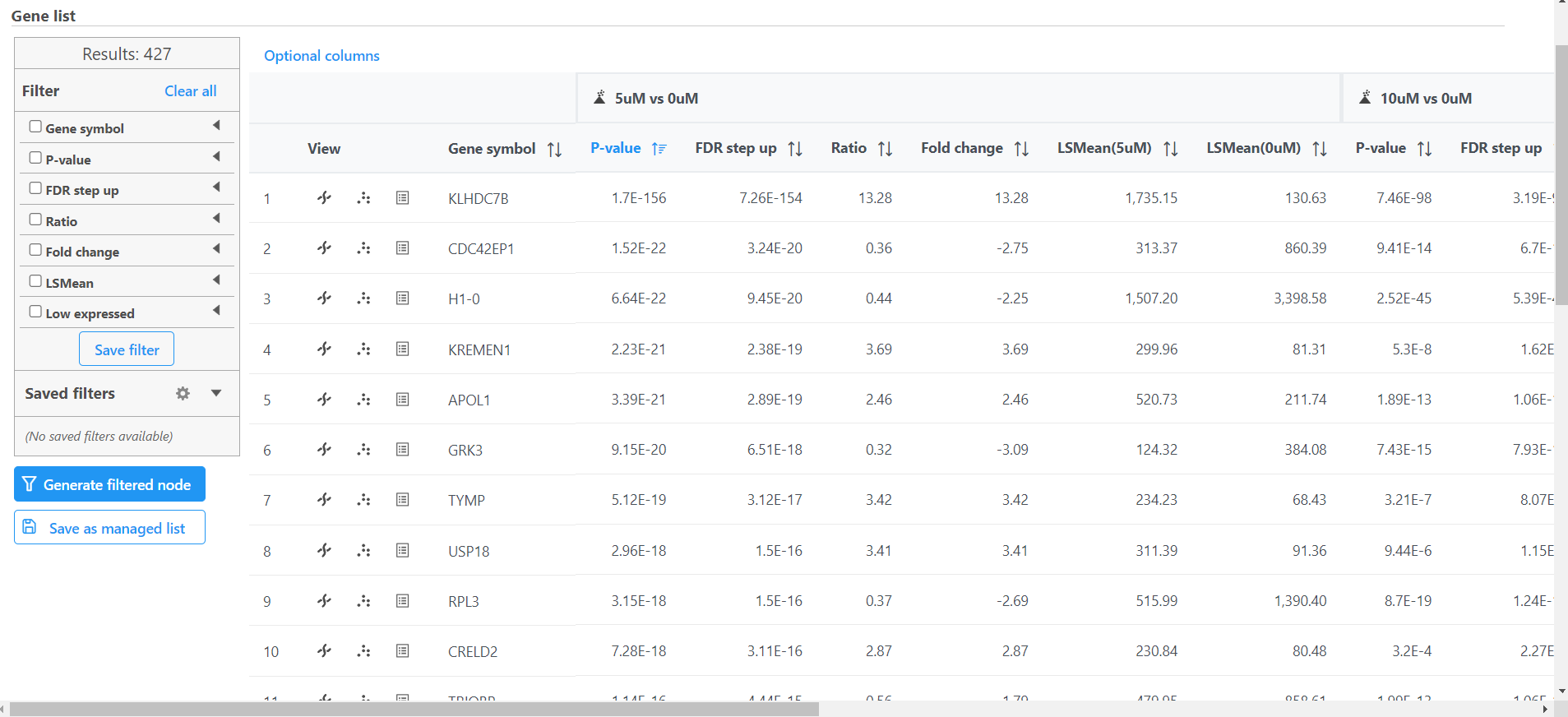
Once we have performed GSA DESeq2 to identify differentially expressed genes, we can create a list of significantly differentially expressed genes using cutoff thresholds.
...
The task report spreadsheet will open showing genes on rows and the results of the GSA DESeq2 on columns (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
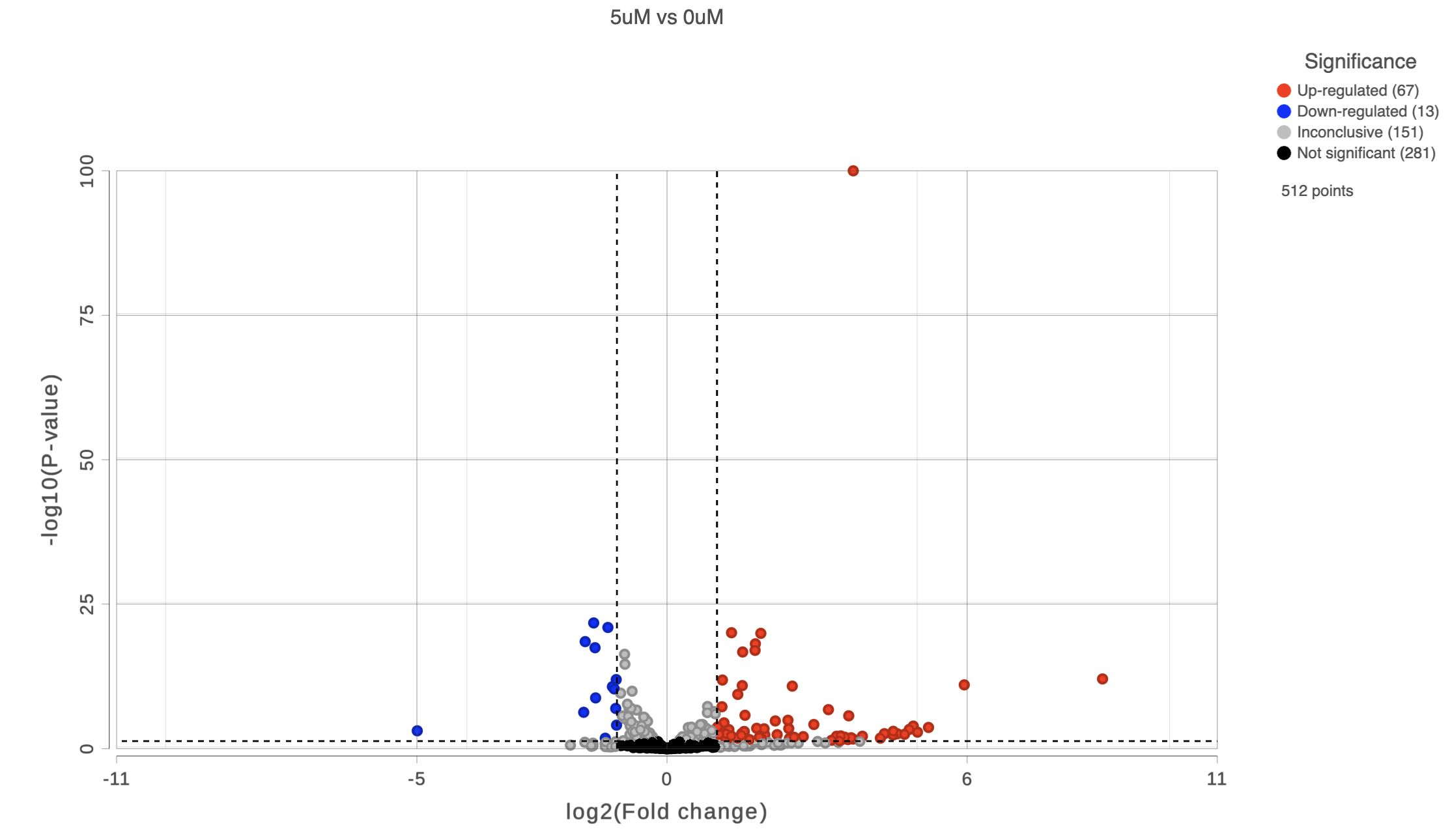
To get a sense of what filtering thresholds to set, we can view a volcano plot for a comparison.
- Select Click next to the 5uM vs. 0uM comparison
A volcano plot will open showing p-value on the y-axis and fold-change on the x-axis (Figure 2).
If the gene labels are on (not shown), click on the plot to turn them off.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Thresholds for the cutoff lines are set using the text boxes on the left-hand side Statistics card (Configuration panel > Configure > Statistics). The default thresholds are 2 are |2| for the X axis and 0.05 for the Y axis.
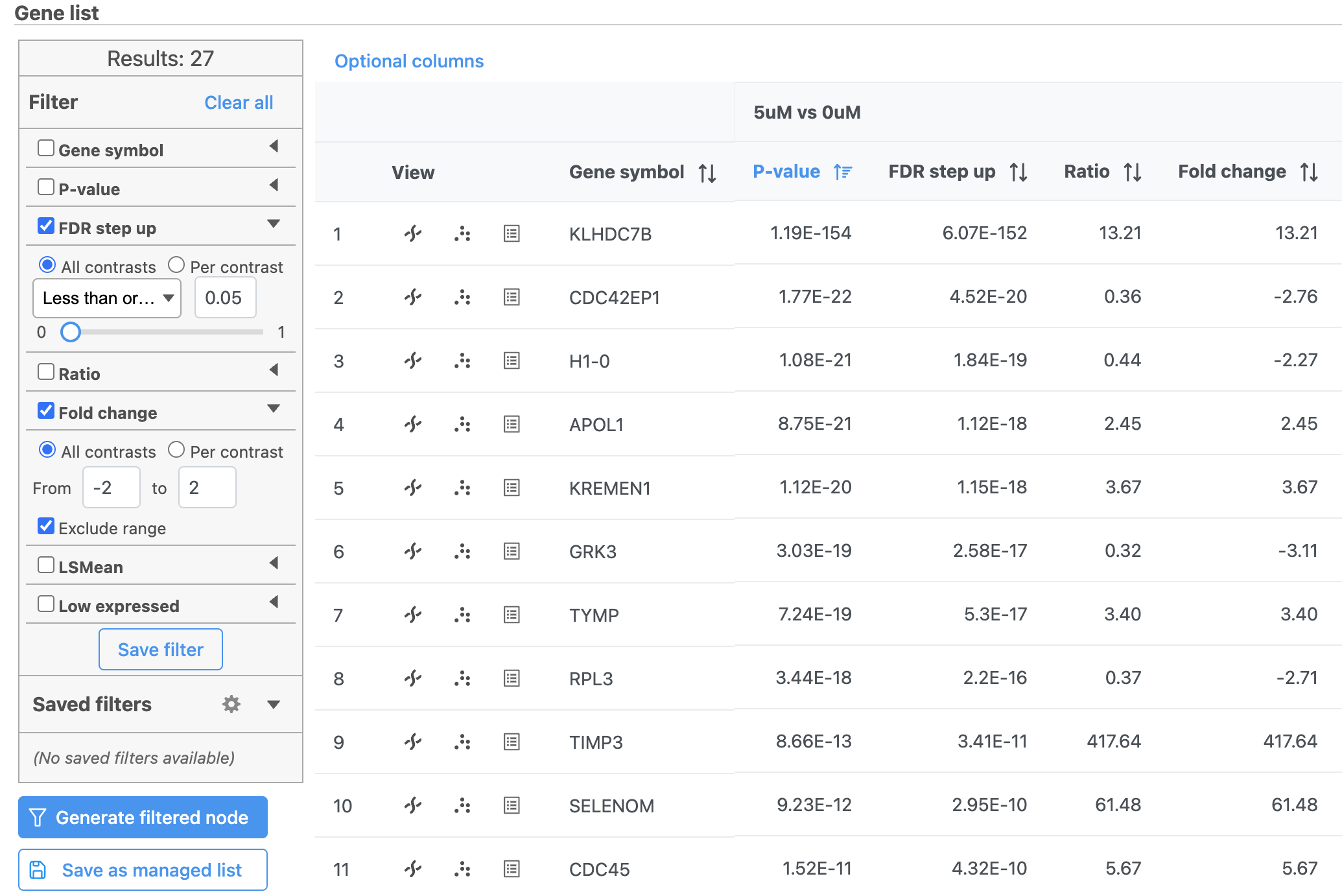
- Select Gene-specific analysis report to return to the task report spreadsheet
- Select Switch to the browser tab showing the DESeq2 report
- Click FDR step up
- Select Click the triangle next to FDR step up to open the FDR step up options
- Select Leave All contrasts selected
- Set the cutoff value to 0.05. Hit Enter.
This will include genes that have a FDR step up value of less than or equal to 0.05 for all three contrasts, 5μM vs. 0μM, 10μM vs. 0μM and 5μM:10μM vs. 0μM. FDR step up is the false discovery rate adjusted p-value used by convention in microarray and next generation sequencing data sets in place of unadjusted p-value.
- Select Click Fold-change
- Select Click the triangle next to Fold-change to open the Fold-change options
- Leave All contrasts selected
- Set to From -2 to 2 with Exclude range selected. Hit Enter.
Note that the number of genes that pass the filter is listed at the top of the filter menu next to Results: and will update to reflect any changes to the filter. Here, 21 27 genes pass the filter (Figure 3).
Depending on your settings, the number may be slightly different.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
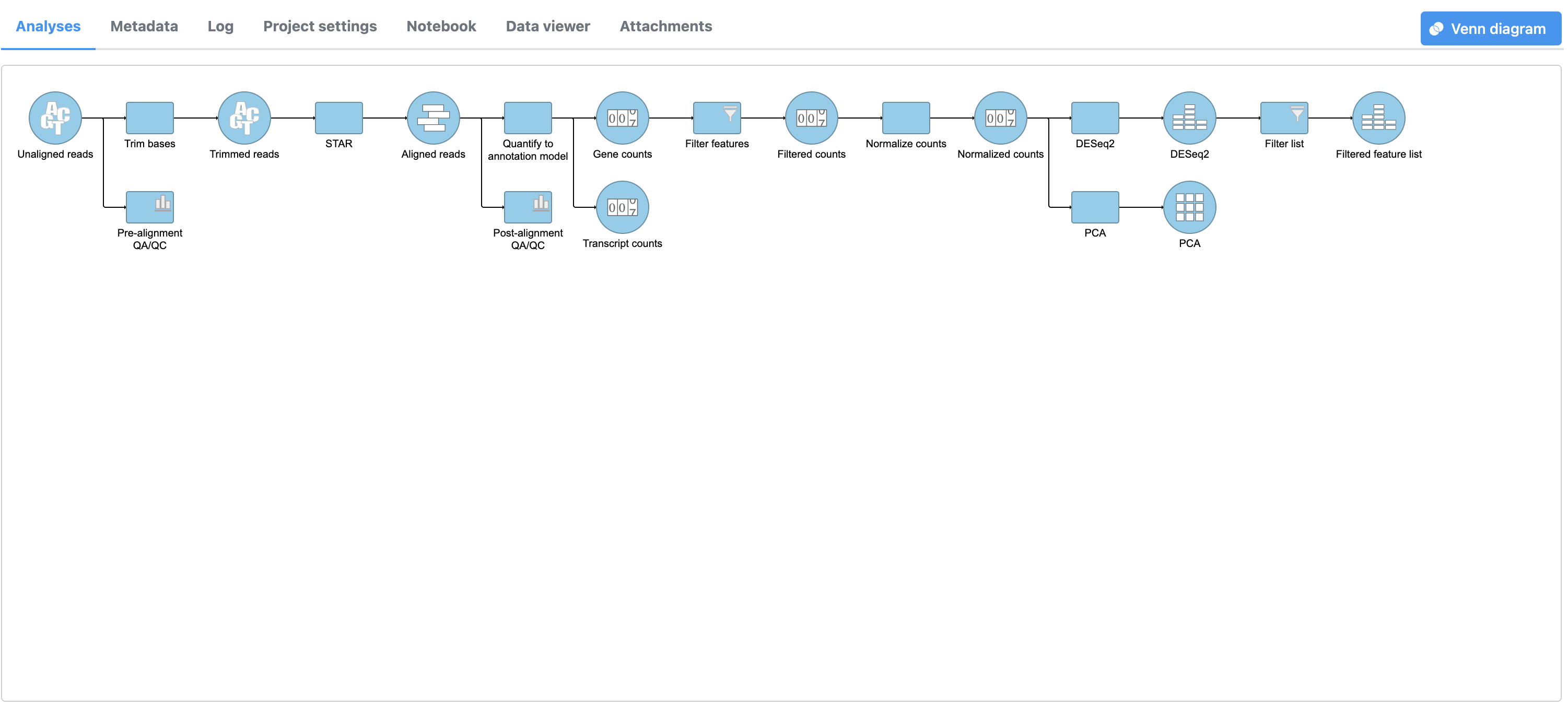
- Select Click to create a data node with only the genes that pass the filter
This creates a Filtered gene analysis Filter list task node and a Feature a Filtered feature list data node (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...