Page History
...
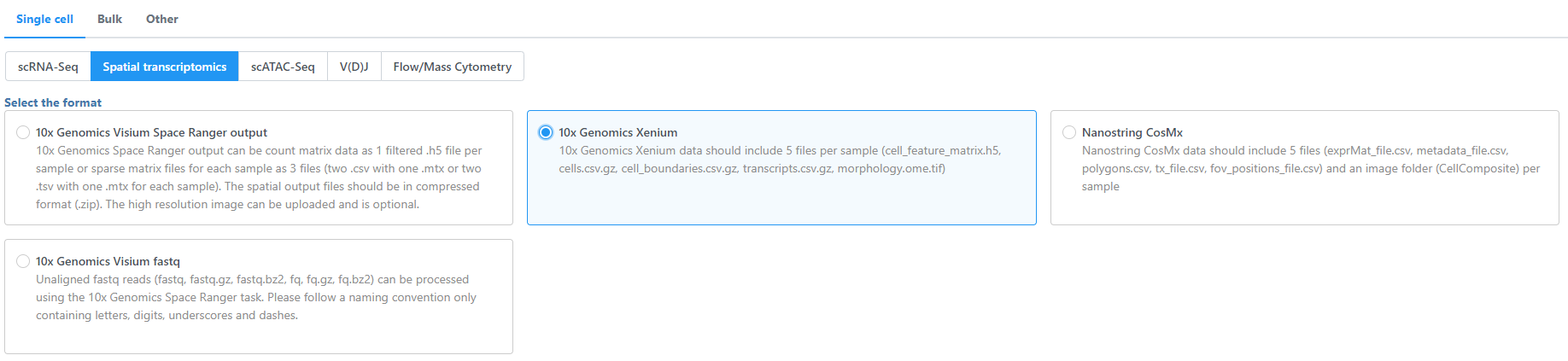
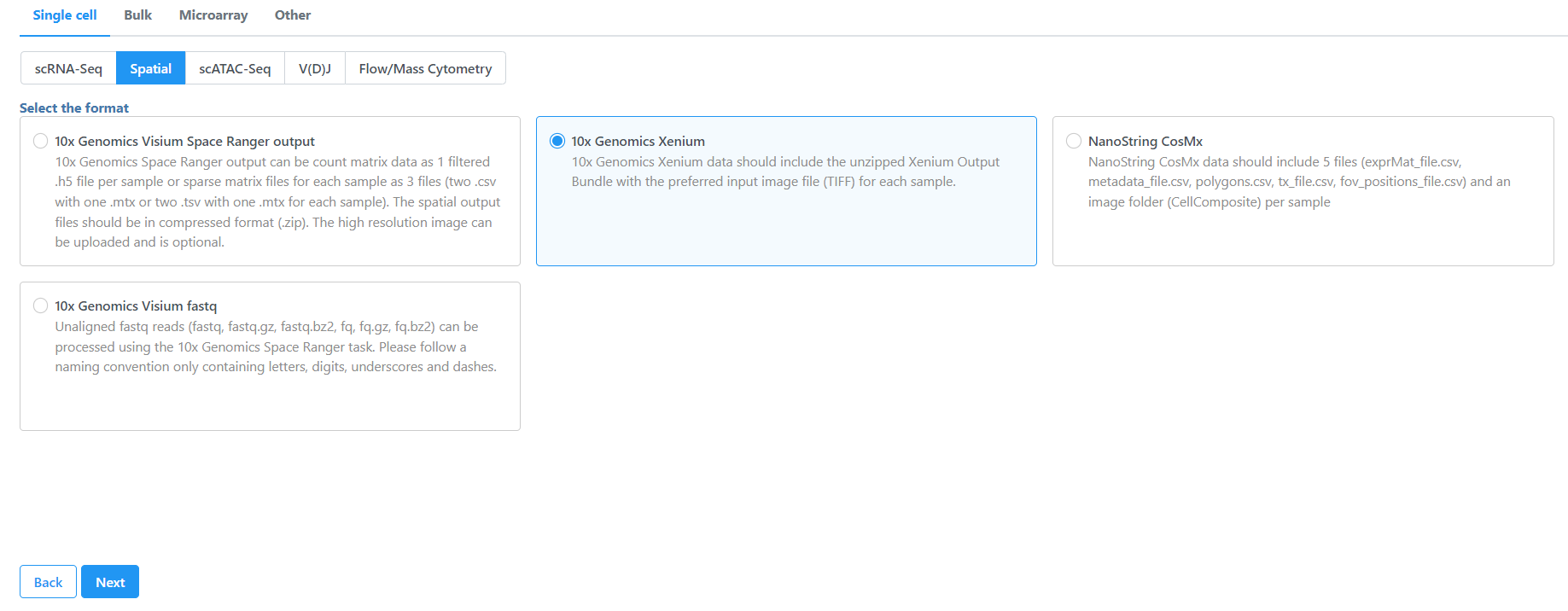
- Navigate the options to select 10x Genomics Xenium Output Bundle as the file format for input. Choose to import 10x Genomics Xenium for your project (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
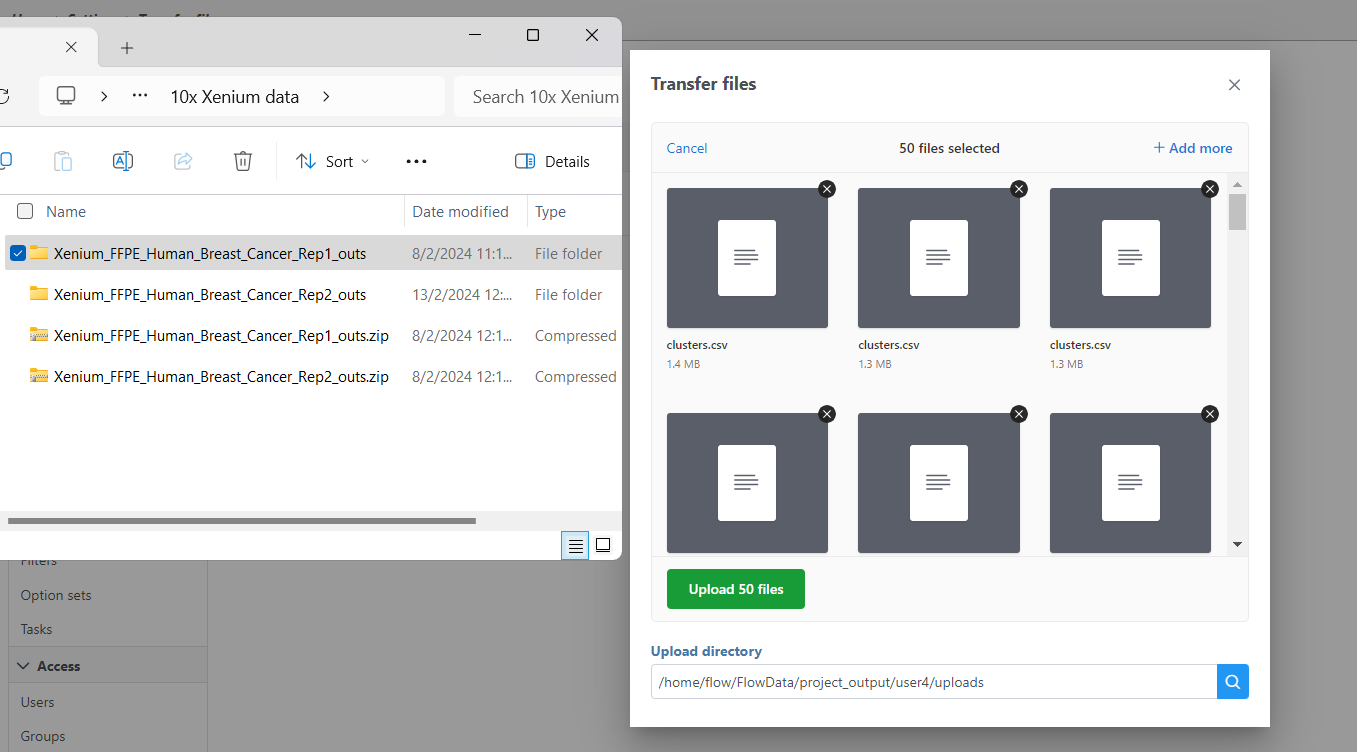
- Click Transfer files on the homepage, under settings, or during import (Figure 3).
...
- .
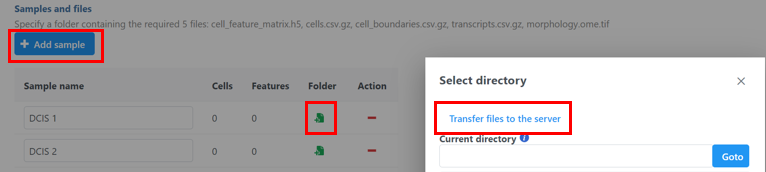
- Click the blue + Add sample button then use the green Add sample button to add each sample's Xenium output bundle folder. If you have not already transferred the folder to the server, this can be done using Transfer files to the server (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Navigate to the appropriate files for each sample (Figure 4You will need to decompress the Xenium Output Bundle zip file before they are uploaded to the server. After decompression, you can drag and drop the entire folder into the Transfer files dialog, all individual files in the folder will be listed in the Transfer files dialog after drag & drop, with no folder structure (Figure 4). The folder structure will be restored after upload is completed.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
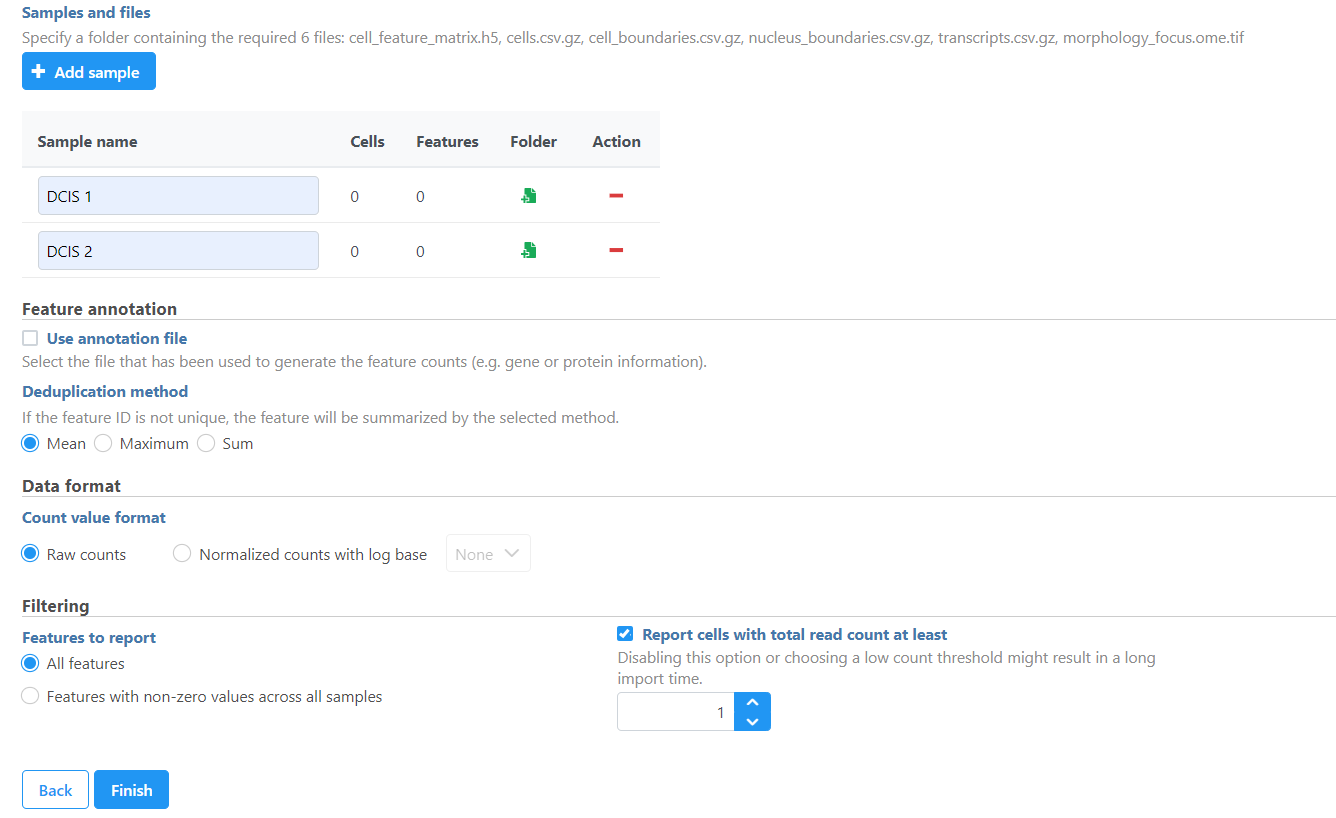
- Once uploaded the folder to the server, navigate to the appropriate folder for each sample using Add sample (Figure 5).
The Xenium output bundle should be included for each sample using Browse (Figure 45). The Each sample requires the whole sample folder can be used, or a folder containing 5 these 6 files (: cell_feature_matrix.h5, cells.csv.gz, cell_boundaries.csv.gz, nucleus_boundaries.csv.gz, transcripts.csv.gz, morphology_focus.ome.tif) can be added per sample. Once added, the Cells and Features values will update. You can choose an annotation file during import that matches what was used to generate the feature count.
Do not limit cells with a total read count since Xenium data is targeted to less features.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
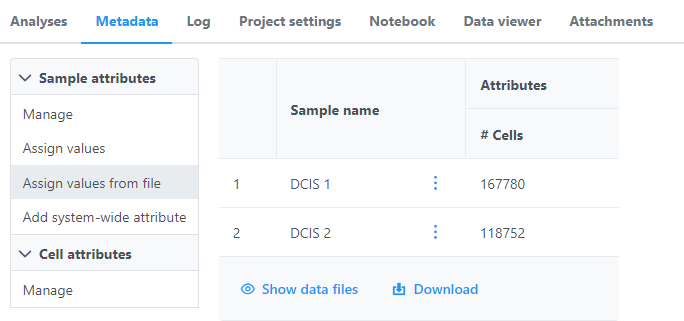
Once the download import completes, the sample table will appear in the Metadata tab, with one row per sample (Figure 56).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The sample table is pre-populated with two one sample attributes: # Cells and Subtype. Sample attributes can be added and edited manually by clicking Manage in the Sample attributes menu on the left. If a new attribute is added, click Assign values to assign samples to different groups. Alternatively, you can use the Assign values from a file option to assign sample attributes using a tab-delimited text file. For more information about sample attributes, see here.
For this tutorial, we do not need to edit or change any sample attributes.
Visualize the annotated image
With samples imported and annotated, we can begin analysis.
- Click Analyses to switch to the Analyses tab
For now, the Analyses tab has only a single node, Single cell counts. As you perform the analysis, additional nodes representing tasks and new data will be created, forming a visual representation of your analysis pipeline.
- Click on the Single cell counts node
A context-sensitive menu will appear on the right-hand side of the pipeline (Figure 9). This menu includes tasks that can be performed on the selected counts data node.
An important step in analyzing single cell RNA-Seq data is to filter out low-quality cells. A few examples of low-quality cells are doublets, cells damaged during cell isolation, or cells with too few counts to be analyzed.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A task node, Single cell QA/QC, is produced. Initially, the node will be semi-transparent to indicate that it has been queued, but not completed. A progress bar will appear on the Single cell QA/QC task node to indicate that the task is running.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The Single cell QA/QC report opens in a new data viewer session. There are interactive violin plots showing the most commonly used quality metrics for each cell from all samples combined (Figure 8). For this data set, there are two relevant plots: the total count per cell and the number of detected genes per cell. Each point on the plots is a cell and the violins illustrate the distribution of values for the y-axis metric. Typically, there is a third plot showing the percentage of mitochondrial counts per cell, but mitochondrial transcripts were not included in the data set by the study authors, so this plot is not informative for this data set.
- Remove the % mitochondrial counts and the extra text box in the bottom right by clicking Remove plot in the top right corner of each plot (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The plots are highly customizable and can be used to explore the quality of cells in different samples.
- Click on Single cell counts in the Get Data icon on the left (Figure 9)
- Click and drag the Sample name attribute onto the Counts plot and drop it onto the X-axis
- Repeat this for the Detected genes plot
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The cells are now separated into different samples along the x-axis (Figure 10)
- Hold Control and left-click to select both plots
- Open the Style icon on the left under Configure
- Under Color, use the slider to reduce the Opacity
- Open the Axis icon on the left
- Adjust the X-rotation on the plots to 90
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Note how both plots were modified at the same time.
Cells can be selected by setting thresholds using the Select & Filter tool. Here, we will select cells based on the total count
- Open Select & Filter under Tools on the left
- Under Criteria, Click Pin histogram to see the distribution of counts
- Set the Counts thresholds to 8000 and 20500
Selected cells will be in blue and deselected cells will be dimmed (Figure 11).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Because this data set was already filtered by the study authors to include only high-quality cells, this count filter is sufficient.
- Click under Filter to include the selected cells
- Click Apply observation filter
- Click the Single cell counts data node in the pipeline preview (Figure 12)
- Click Select
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new task, Filter counts, is added to the Analyses tab. This task produces a new Filter counts data node (Figure 13).
- Click on the Glioma (multi-sample) project name at the top to go back to the Analyses tab
- Your browser may warn you that any unsaved changes to the data viewer session will be lost. Ignore this message and proceed to the Analyses tab
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Most tasks can be queued up on data nodes that have not yet been generated, so you can wait for filtering step to complete, or proceed to the next section.
Filtering genes in single cell RNA-Seq data
A common task in bulk and single-cell RNA-Seq analysis is to filter the data to include only informative genes. Because there is no gold standard for what makes a gene informative or not, ideal gene filtering criteria depends on your experimental design and research question. Thus, Partek Flow has a wide variety of flexible filtering options.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
There are four categories of filter available - noise reduction, statistics based, feature metadata, and feature list (Figure 15).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The noise reduction filter allows you to exclude genes considered background noise based on a variety of criteria. The statistics based filter is useful for focusing on a certain number or percentile of genes based on a variety of metrics, such as variance. The feature list filter allows you to filter your data set to include or exclude particular genes.
We will use a noise reduction filter to exclude genes that are not expressed by any cell in the data set but were included in the matrix file.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
This produces a Filtered counts data node. This will be the starting point for the next stage of analysis - identifying cell types in the data using the interactive t-SNE plot.
Normalizing single cell RNA-Seq data
We are omitting normalization in this tutorial because the data has already been normalized.
The tutorial data set is taken from a published study and has already been normalized using TPM (Transcripts per million), which normalizes for the length of feature and total reads, and transformed as log2(TPM/10+1). This normalization and transformation scheme can be performed in Partek Flow, along with other commonly used RNA-Seq data normalization methods.
For more information on normalizing data in Partek Flow, please see the Normalize counts section of the user manual.
Cell attributes are found under Sample attributes and can be added by publishing cell attributes to a project.
For this tutorial, we do not need to edit or change sample attributes.
| Page Turner | ||
|---|---|---|
|
...