Page History
Dot plot is a statistical chart, typically used to gene expression (RNA-seq or microarray data), with dots representing samples and y-axis showing expression level on an appropriate scale.Dot plot is a statistical chartspecial 2D scatterplot with X-axis is a categorical variable and Y-axis is a numerical variable, typically used to visualize gene expression (RNA-seq or microarray datascRNA-seq), with dots representing observations (samples or cells), X-axis showing treatment groups and y-axis showing expression level on an appropriate scale.
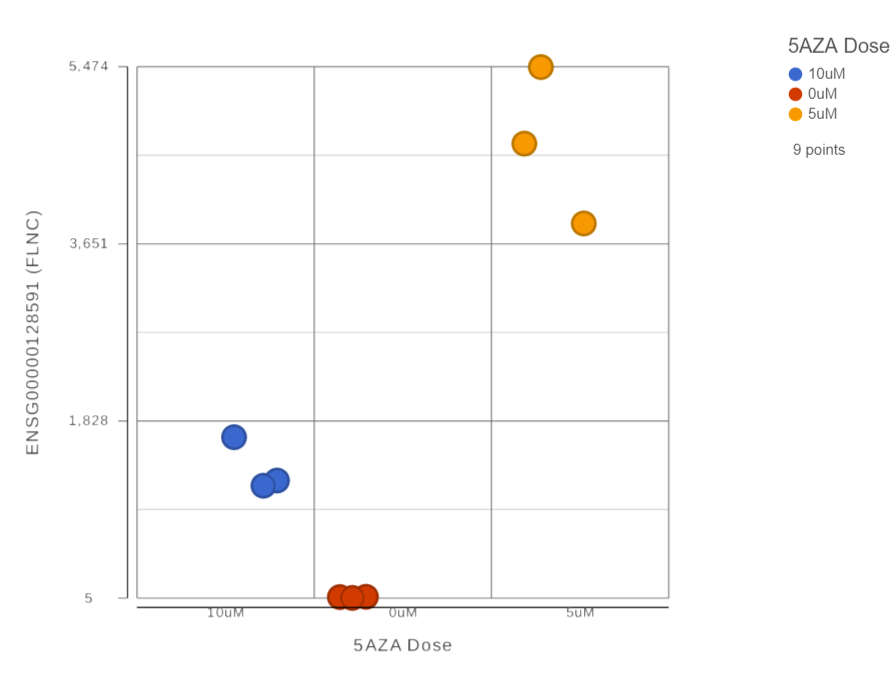
In Partek® Flow®, dot plots are available through a Feature list node, by selecting differential analysis report data node, select the dot plot icon ( ) in the View column to invoke the plot in data viewer. The plot will be displayed in a new browser tab (an example plot is on Figure 1). Figure 1:
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The chart title is based on the genomic feature (e.g. gene or transcript) that the plot was invoked on. The y-axis is scaled automatically, based on the range of the data, and the units correspond to the input units of the parent Feature list node, i.e. if the data were normalized using transcripts per million (TPM), the y-axis will be in TPM-normalised counts. Dots represent observations (e.g. samples or cells). Hovering the cursor over a sample invokes a popup balloon message shows sample ID and the respective expression value. The legend is in the upper right corner and is based on the data attribute specified under the Color by option (on the left).
Plot controls are in the panel on the left. The <Previous and Next> buttons enable you to switch between genomic features as they appear in the parent Feature list. The Group by option defines the grouping on the x-axis, with the drop-down list containing the data attributes as present in the Data tab. As previously mentioned, the dot colors are controlled by the Color by option. If you want to change a color, select the Customize colors hyperlink. The resulting dialog (Figure 2) will enable you to replace an existing color by a color of your choice (click on the arrow head to invoke the color mixer) or add more colors (Add color).
...
Figure 4: Save image dialog (default settings)
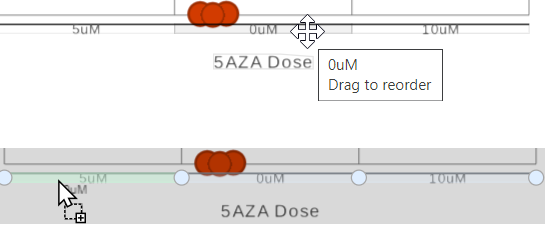
Use the Group order section of the control panel to change the order of the groups on the x-axis. Simply drag and drop a group label to a new position.
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
The order of the group on the X-axis by default is based on the order in the attribute management page in the Data tab. However, you can manually click on the name of the group on X-axis, drag and drop to change the order (Figure 2)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
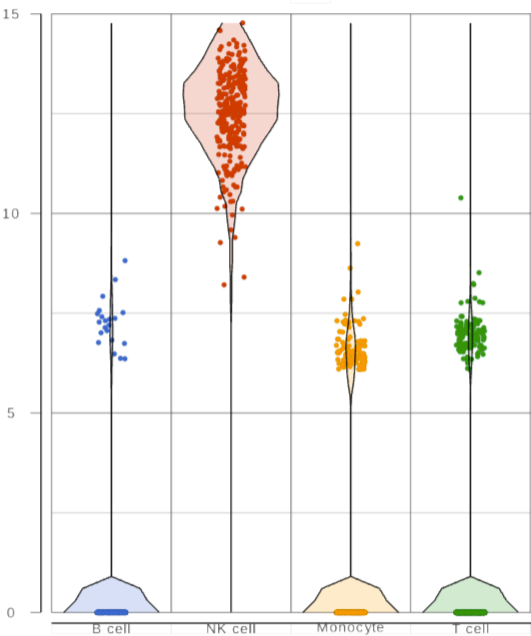
When you have a lot of observations to display, you can choose to add box-whiskers and/or violin plot on the graph (Figure 3), by turning on/off the options to display the different type of plots in Style card from the configuration panel.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
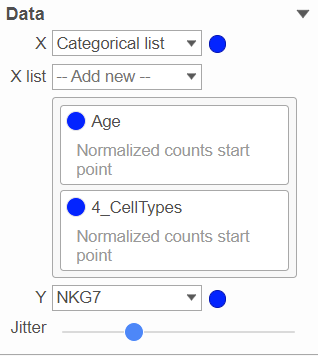
You can have multiple categorical attributes on an axis, e.g. X-axis represent both cell type and age group (Figure 4). Click on the attribute to drag to change the order of the attributes
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
ghjkl
Click the Export image button to save a PNG, SVG, or PDF image to your computer.
Click the To notebook button to send the image to a page in the Notebook.
ghjkl
ghjkl
ghjkl
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|