Page History
...
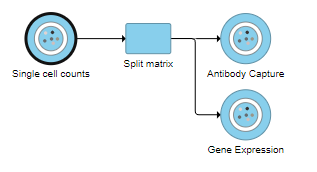
Split matrix can be invoked on any counts data node with more than one feature type. For example, a CITE-Seq experiment would have Gene Expression counts and Antibody Capture counts in the single cell counts data node. Datasets generated by 10X Genomics' Feature Barcoding experiments also utilize this task to split different feature measurements for downstream analysis.
There are no parameters to configure, to run:
...
The Split matrix task will run and generate output data nodes for each of the feature types. For example, if there are Antibody Capture and Gene Expression feature types in the input, Split matrix will generate two data nodes (Figure 1). Every sample is included in both matrices.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...
Overview
Content Tools