Page History
...
The tutorial data set includes 9 samples equally divided into 3 treatment groups. Sequencing was performed by an Illumina HiSeq (paired-end reads), but the workflow can be easily adapted for data generated by other sequencers. Each sample has 2 fastq files for a total of 18 fastq files with the combined size of 6.17 GB.
This tutorial will use two sets of samples: human brain reference (HBR) samples and universal human reference (UHR) samples (five each). Data was generated using an Ion Torrent® sequencer (single-end chemistry), but the workflow can be easily adapted for data generated by sequencers from other manufacturers. The project consists of 10 fastq files with the combined size of 1.4 GB.
To .
You can obtain the tutorial data set , select through Partek Flow.
- Click your avatar
...
- Click Settings in the drop-down menu (Figure 1)
...
| Numbered figure captions | ||
|---|---|---|
|
...
| |||
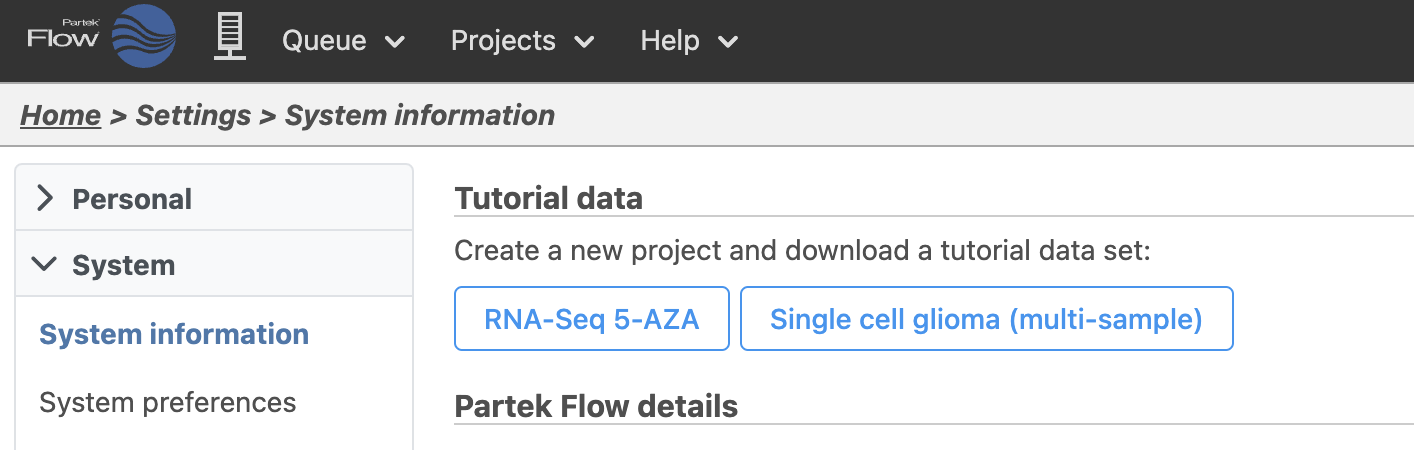
At the top of the System information page, there is a section labeled Download tutorial data (Figure 2)
...
.
...
| Numbered figure captions | ||
|---|---|---|
|
...
| ||
|
- Click RNA-Seq 5-AZA to download the tutorial data set
A new project will be created and you will be directed to
...
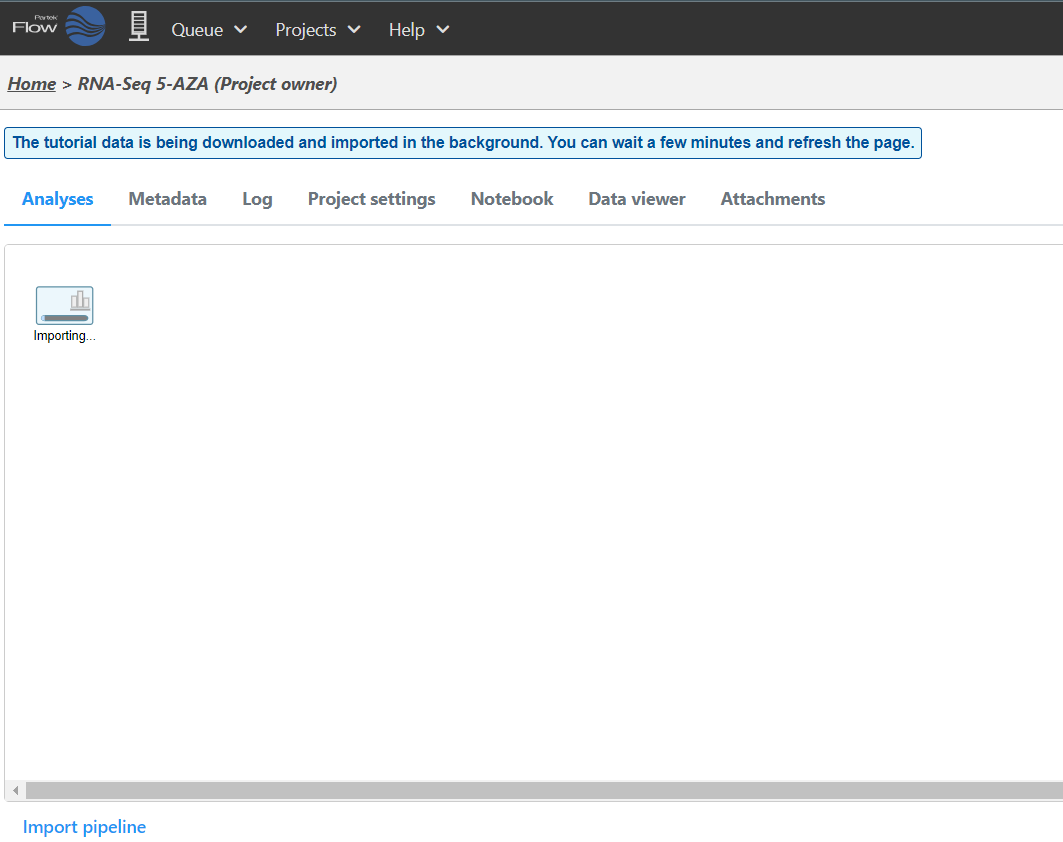
the Analyses Tab. The data will be downloaded automatically (Figure 3) and imported into your project.
...
Because this is a tutorial project, there is no need to click on
...
Add data as it will be done automatically.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
At first the project is empty, but the file download will start automatically in the background. You can wait a few minutes
...
then refresh
...
your browser or you can monitor the download progress
...
using the Queue.
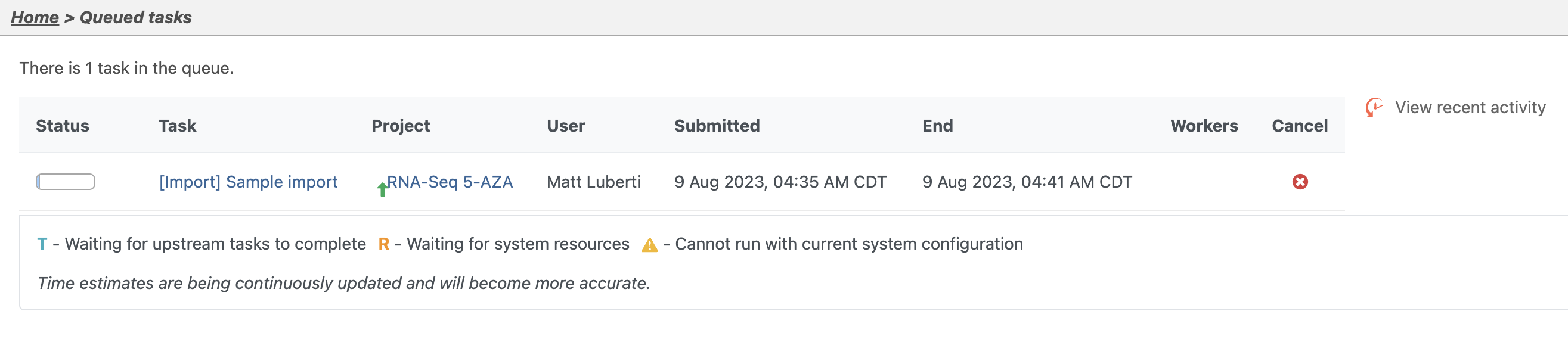
- Click Queue
- Click View Queued Tasks in the drop-down menu
The Queued tasks page will open (Figure 4)
...
.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click Projects
- Click RNA-Seq 5-AZA in the drop-down menu
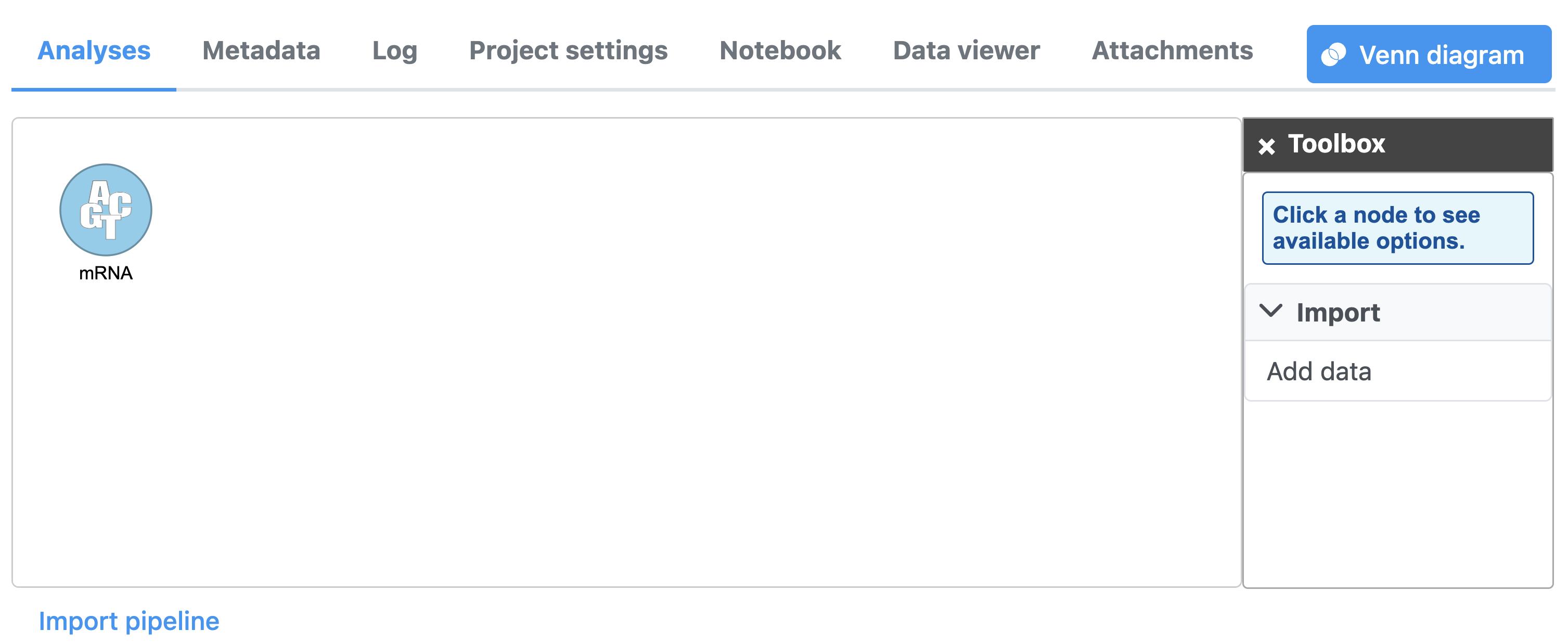
The Analyses tab will open (Figure 5). If you download has completed, you will see a blue circle titled mRNA.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Once the download completes, the sample table will appear in the Data tab (Figure 5).
...
the Metadata tab.
- Click the Metadata tab
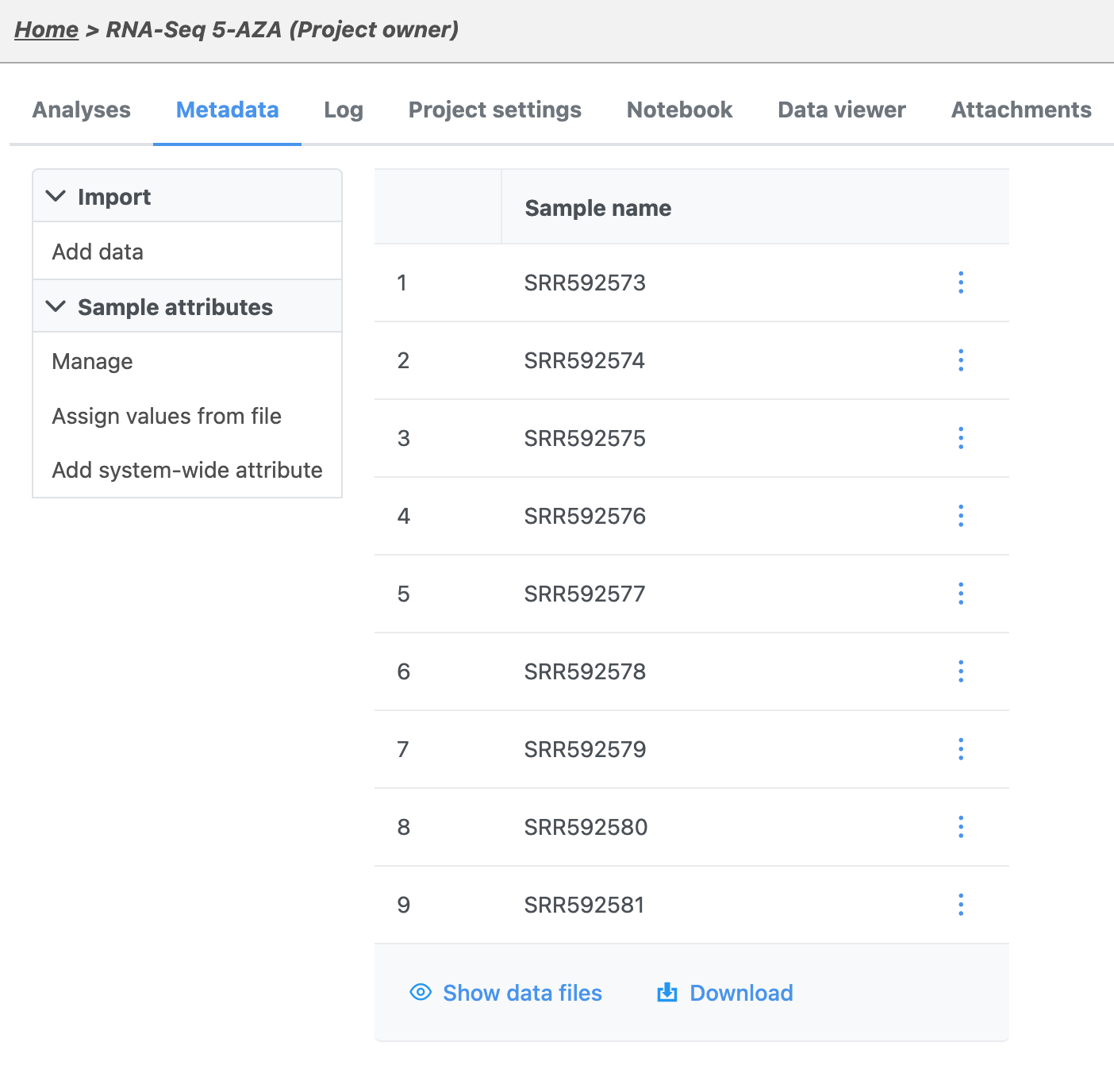
The Metadata tab includes the sample table with the names of each imported sample (Figure 6).
| Numbered figure captions | ||
|---|---|---|
|
...
|
...
| |||
In the next section of the tutorial, we will add a sample attribute that indicates the treatment group of each sample.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...