Page History
...
- Open the filtering gene list and target spreadsheets
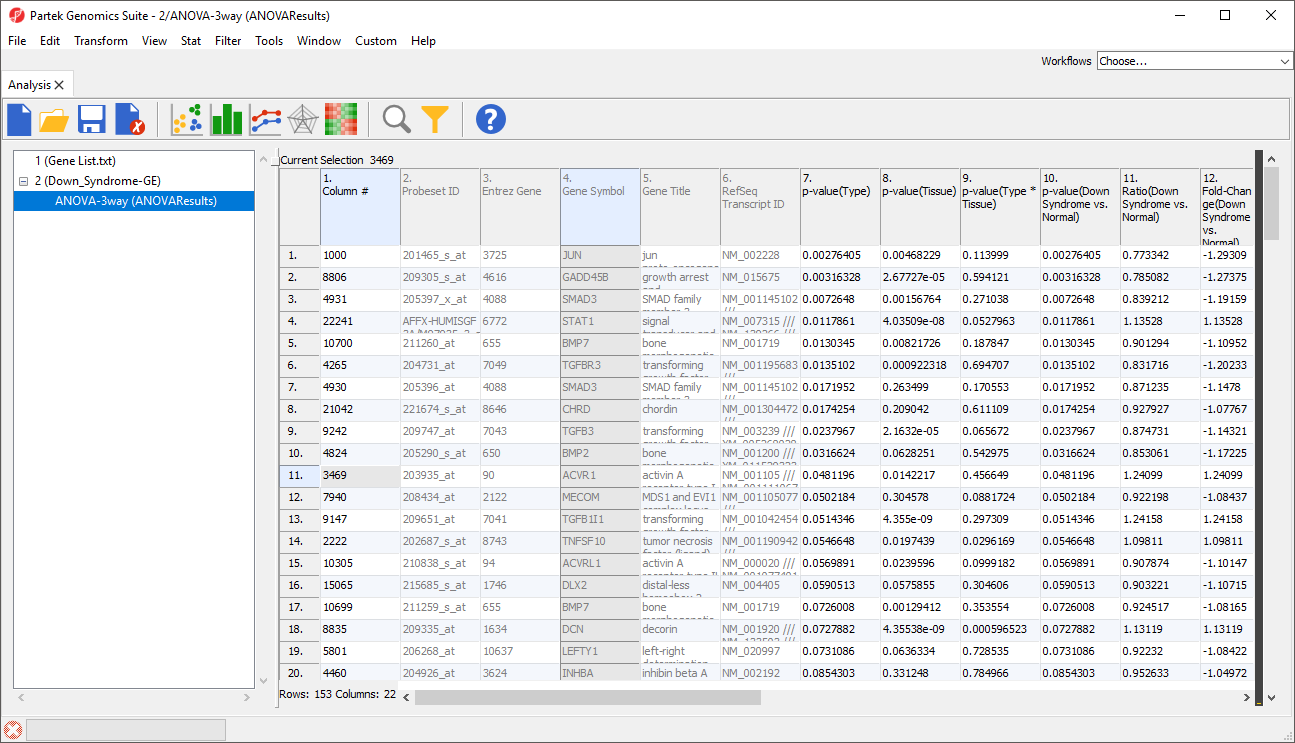
- Select the target spreadsheet in the spreadsheet tree, in this example, genes are on rows in ANOVA result spreadsheet
- Select Filter from the main toolbar
- Select Filter Rows Based on a List... from Filter Rows (Figure 1)
...
The target spreadsheet will display the filtered rows (Figure 3). Note that the number of rows has gone from 22,283 prior to filtering (Figure 1) to 153 after filtering (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
The new spreadsheet will open. If you want to use the list new spreadsheet again in the future, be sure to save it as the new spreadsheet is a temporary file.
Descriptive Statistics
There are numerous descriptive statistics available in Partek Genomics Suite.
- Select Stat from the main toolbar
- Select either Descriptive or Correlate to show available options
Principal Component Analysis is located in a different menu.
- Select Tools from the main toolbar
- Select Discover
- Select Principal Component Analysis
Applying Multiple Test Correction
...
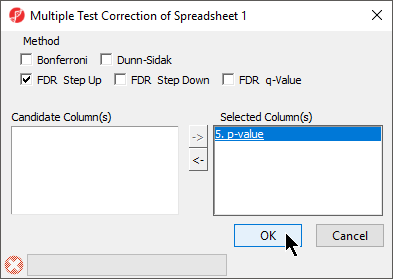
- Select Stat from the main toolbar
- Select Multiple Test
- Select Multiple Test Corrections to launch a dialog with available options (Figure 1)4), it will add corrected p-value column(s) to the right of the selected p-value column(s)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
If you have imported numerical data associated with genes (like p-values or fold-changes), you can visualize these values in the Genome Browser once an annotation file has been addedis associated to the spreadsheet, and there is genomic location information in the annotation file.
- Right-click on a row header in the imported gene list spreadsheet
- Select Browse to location
If the annotations have been configured properly, you should see a track Regions track for the first column of numerical data, a cytoband track, and an annotation track. You can also add another track to display a second column of numerical data.
...
- Select Regions in the track preferences panel to edit it
- Select the other numerical column in the Bar height by drop-down menu
Clustering
If the data is suitable for clustering, access For a gene list with expression values on each sample, clustering can be performed. Access the clustering function through the toolbar, not form from a workflow. The workflow implementation assumes implementations assume that the data to be clustered are found on a parent spreadsheet and the list of genes is in a child spreadsheet. Because the data to be clustered is all on one spreadsheet, access hierarchical clustering by selecting Tools from the main toolbar then Discover then Hierarchical Clustering. Consider transposing the spreadsheet if samples are on columns and genes are on rows as Hierarchical Clustering will assume
- Select Tools form the main toolbar
- Select Discover then Hierarchical Clustering
Hierarchical Clustering assumes that samples are rows and genes are columns so consider transposing your data if this is not the case. If you have only have one column or one row of data, cluster only on the dimension with multiple entries categories by deselecting either deselecting either Rows or Columns from What to Cluster or consider using an intensity plot insteadCluster in the Hierarchical Clustering dialog.
| Page Turner | ||
|---|---|---|
|
...