Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
This white paper explains some basic concepts related to alignment and mapping quantification tasks in Partek® Genomics Suite™ (PGS) v6.6 under the RNA-seq workflow. Flow. The term “alignment” describes the process of finding the position of a sequencing read on the reference genome, while “mapping” quantification refers to assigning already aligned reads to transcripts ; it is also called quantification.based on specified transcripts/gene annotation.
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Number of Reads and Alignments
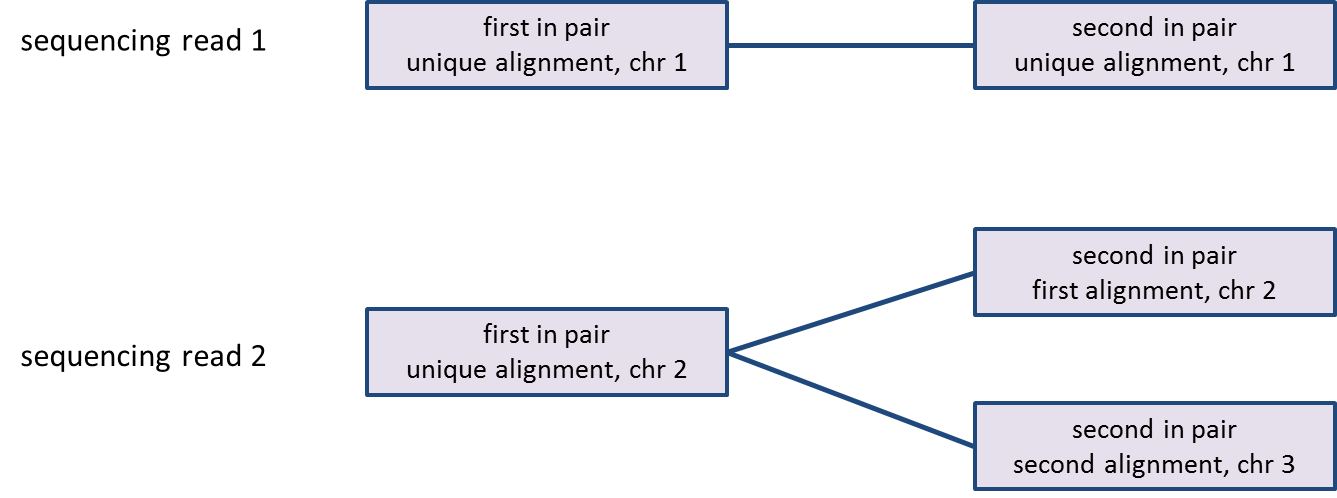
PGS imports all the sequencing Aligned data node contains reads that have been aligned, counting each read once even if it has multiple alignments. Processing of paired-end reads needs further elaboration: a paired-end read will be counted as one read only if both ends align to the genome, no matter how many alignments each end has (Figure 1). If one end of a paired-end read is not aligned, the read will be discarded (i.e., not imported).considered unaligned and will be discarded for the downstream analysis.
As reads can be aligned to more than one location, the number of alignments may be greater than the number of reads; since some reads may be unaligned, the number of alignments may be less than the number of reads in the bam/sam file (Figure 1).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
PGS Partek Flow shows the number of alignments per sample in the “parent” spreadsheet of a RNA-seq project, while the alignments and reads per sample are reported in the mapping_summary spreadsheet. The alignment_counts spreadsheet contains the number of alignments per read in each sample and it can be invoked through the QA/QC section of the RNA-seq workflowalignments as well as the aligned reads percentage per sample in the post alignment QC report.
Quantification: Assigning Reads to Transcripts
This step maps the aligned reads to transcripts using a modified E/M algorithm; detailed information about this algorithm can be found in the white paper RNA-Seq methods in the tutorial pageSeq quantification.
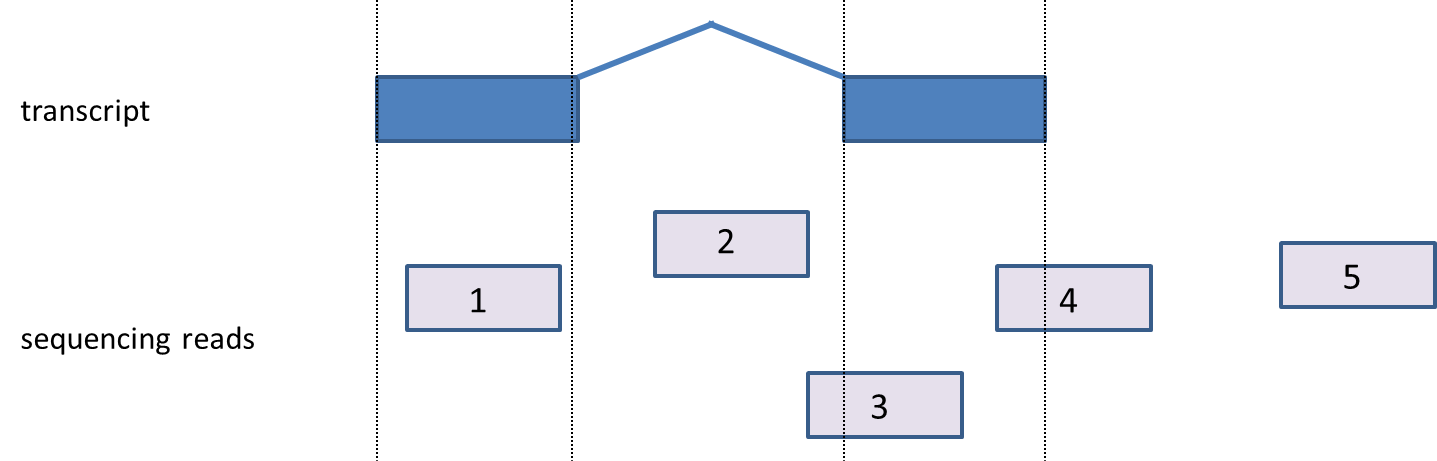
Given a list of transcripts, each read can be mapped to one of four classes: exonic, partially overlaps exon, intronic, or between genes (Figure 2). Generally, “exonic” means the read came from a sequence present in the mature mRNA, while “intronic” means the read overlaps a gene, but aligns to the portion of the sequence not present in the mature mRNA. Finally, “between genes” reads came from a region outside of any gene. Detailed definitions of all four types can be found below.
...
- Exonic: A read is labeled exonic if any one of its alignments is completely contained within the respective exon as defined by the database (read 1) ,i.e., even if there is a single base shift of the read relative to the exon, the read will not be called exonic but will fall in the category ‘partially overlaps exon’). If the alignments are strandspecificstrand specific, then the strand of the alignment must also agree with the strand of the transcript.
- Partially overlaps exon: A read is assigned to partially overlap an exon if any of its alignments overlaps an exon, but at least partly (one base-pair or more) maps out of the exon (read 3, read 4).
- Intronic: A read is labeled intronic if any one of its alignments maps completely within an intron (read 2), but none of the alignments are exonic (either fully or in part). If the alignments are strand-specific, then the strand of the alignment must also agree with the strand of the transcript.
- Reads between genes: A read is labeled ‘between genes’ if none of its alignments overlap a gene (read 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
RPKM Scaling
Standard output of mapping performed by the quantification step includes raw read counts and scaled read counts for every gene and transcript for each sample. The scaling method currently applied is reads per kilo-base of exon model per million mapped reads (RPKM) (Mortazavi et al. Nat Methods 2008). It scales the abundance estimates using exon length and millions of mapped reads and is calculated according to the formula below.
FORMULA
For example, suppose a transcript has 50 reads that map to it, with an exon transcript length of 7000 bp ( = 7.0 kbp), and there are 5,000,000 reads in that sample. The RPKM value is:
500/(7.000) × (1,000,000/5,000,000) = 14.28 RPKM reads
The exon length used in the formula is the sum of constituent exons, not the distance from the transcription start to the transcription stop. Moreover, as stated above, PGS uses the total number of mapped reads (not alignments!) for the RPKM calculation. In the other words, a paired-end read is counted as one read.
When adding up all of the RPKM reads for all the transcripts in a gene, they do not necessarily equal the gene level RPKM value. Only reads mapping to a transcript as exonic reads (as defined previously) will be counted as reads for that transcript. Reads partially overlap an exon or in the intronic regions are counted for gene level summarization .
During the quantification step, all aligned reads presented in the BAM file are used; no matter they are uniquely aligned or multiply aligned. If a read is uniquely aligned, then it contributes 1 count to the respective transcript. On the other hand, if a read aligns to multiple locations, then the algorithm will divide that read proportionally amongst the transcripts, depending on how likely the read maps to one or the other.
Read Compatibility
A read will be assigned to a transcript only when it is compatible. A compatible read is a read that fits the transcript model from the chosen annotation database. Compatible reads must be an exonic read (fully mapped to exon); however not all the exonic reads are compatible with a transcript, e.g., in paired end reads, both end reads have to be exonic as well as they both have to map to the same transcript; if they mapped to two different transcript or different chromosome, they are not compatible with a transcript.
Reads that are partially mapped exon reads, intronic reads, intergenic reads are incompatible with any transcript. Incompatible reads do not contribute to gene or transcript level read counts. Several single-end and paired-end scenarios will be discussed in the following sections.
In addition, the implementation of the compatibility rules has been changed between PGS releases 6.5 and 6.6 thus leading to possible differences in the read counts between the two versions. The major difference is that 6.6 is more strict: 6.5 would count a paired-end read as compatible if it had any alignments that are compatible as opposed to 6.6 which requires both first-in-pair and second-in-pair to be compatible.
Single-End Scenarios
A single-end read that is considered “compatible” would be any read that overlaps the exon 100% as described above in the paragraph on “exonic” reads. For compatible reads, PGS gives raw read counts and RPKM (scaled) counts, while incompatible reads are presented by RPKM counts only (“incompatible RPKM”) and are given in the transcripts spreadsheet (Figure 3). When calculating the RPKM values for incompatible reads, PGS still uses the exon length, as previously described.
If a single-end read includes an exon to exon junction, the region skipped in the read must be consistent with an intron in that transcript. If it is not, the read will be incompatible.
For example, with an alignment that begins at base 100 and has a cigar score of 5M 10N; 5M, the M means alignment Match and the N means skipped bases (junction). To be compatible, bases 100-104 must be exonic, bases 105-115 must be intronic and bases 116-120 must be exonic.
Compatible reads with junction are reported in a separate block of columns in the transcripts spreadsheet (“junction RPKM”).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
Paired-End Data Scenarios
...
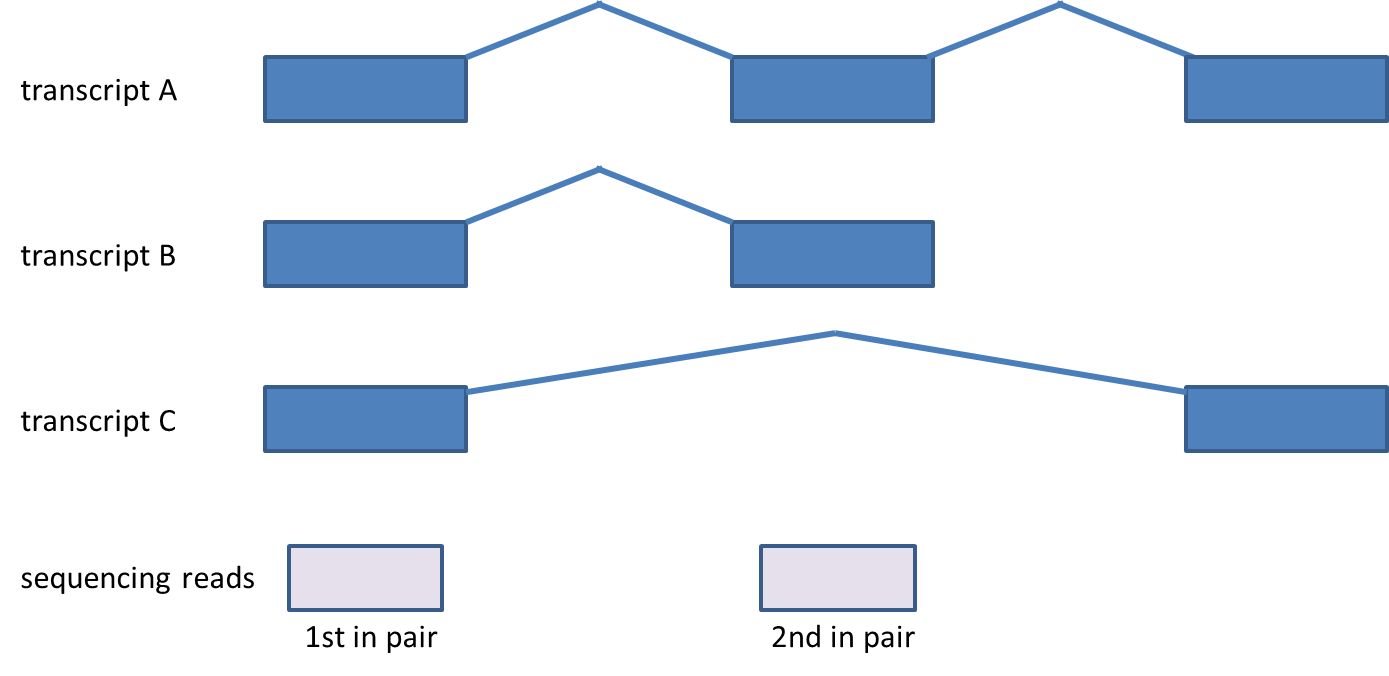
Considering genes with multiple transcripts, a read can be both counted as compatible for some transcripts, as well as counted incompatible for other transcripts of the same gene (Figure 43). Please note that this concept holds for single-end reads as well.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
Furthermore, all the reads that have at least one alignment contribute to the denominator of the RPKM (millions of _mapped reads_ per sample). Similar to that, the denominator of the RPKM contains all the mapped reads, regardless of whether or not a read is compatible with any transcript.
With respect to the gene-level summarization, user can decide whether or not to consider intronic reads (both single- and paired-end) as compatible with the gene (Figure 5). In the latter case, the entire gene is basically treated as one giant exon. In the case that one end of a paired-end read falls within a gene, and the other end does not, the read will not be included in the gene-level summarization.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
total number of aligned reads.
Unexplained regions
The unexplained regions portion of quantification considers any read that is considered “not compatible” with all transcripts. It is basically a 3 step process:
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
...
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...