...
- Click the Merged counts data node
- Click Exploratory analysis in the toolbox
- Click PCA
- Click Finish to run the PCA with default settings (Figure ?1)
| Numbered figure captions |
|---|
| SubtitleText | Run PCA with default settings |
|---|
| AnchorName | PCA task set up |
|---|
|
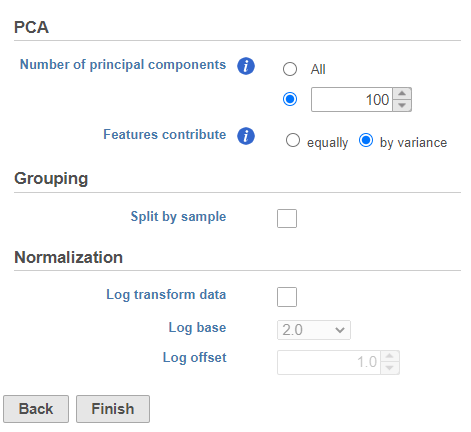 Image Removed Image Removed Image Added Image Added
|
A 388pxA PCA task node will be added to the pipeline under the Analyses tab and a circular PCA output data node will be produced (Figure ?2).
| Numbered figure captions |
|---|
| SubtitleText | PCA task run on the merged counts data node |
|---|
| AnchorName | PCA output node |
|---|
|
|
...
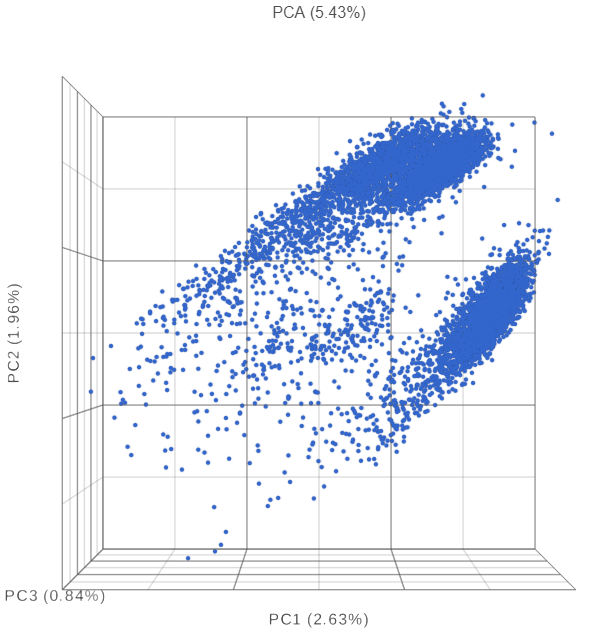
The PCA plot will open in a new data viewer session. A 3D scatterplot will be displayed on the canvas (Figure ?3).
| Numbered figure captions |
|---|
| SubtitleText | Each dot is a different cell. Cells are clustered based on how similar their expression profile is across the combined mRNA and protein data |
|---|
| AnchorName | PCA merged counts |
|---|
|
|
- Click and drag the Scree plot from the Available plots card New plot under Setup on the left onto the canvas
- Drop it over the Replace option (Figure ?4)
| Numbered figure captions |
|---|
| SubtitleText | Click and drag the Scree plot to replace the PCA plot on the canvas |
|---|
| AnchorName | Replace PCA with Scree plot |
|---|
|
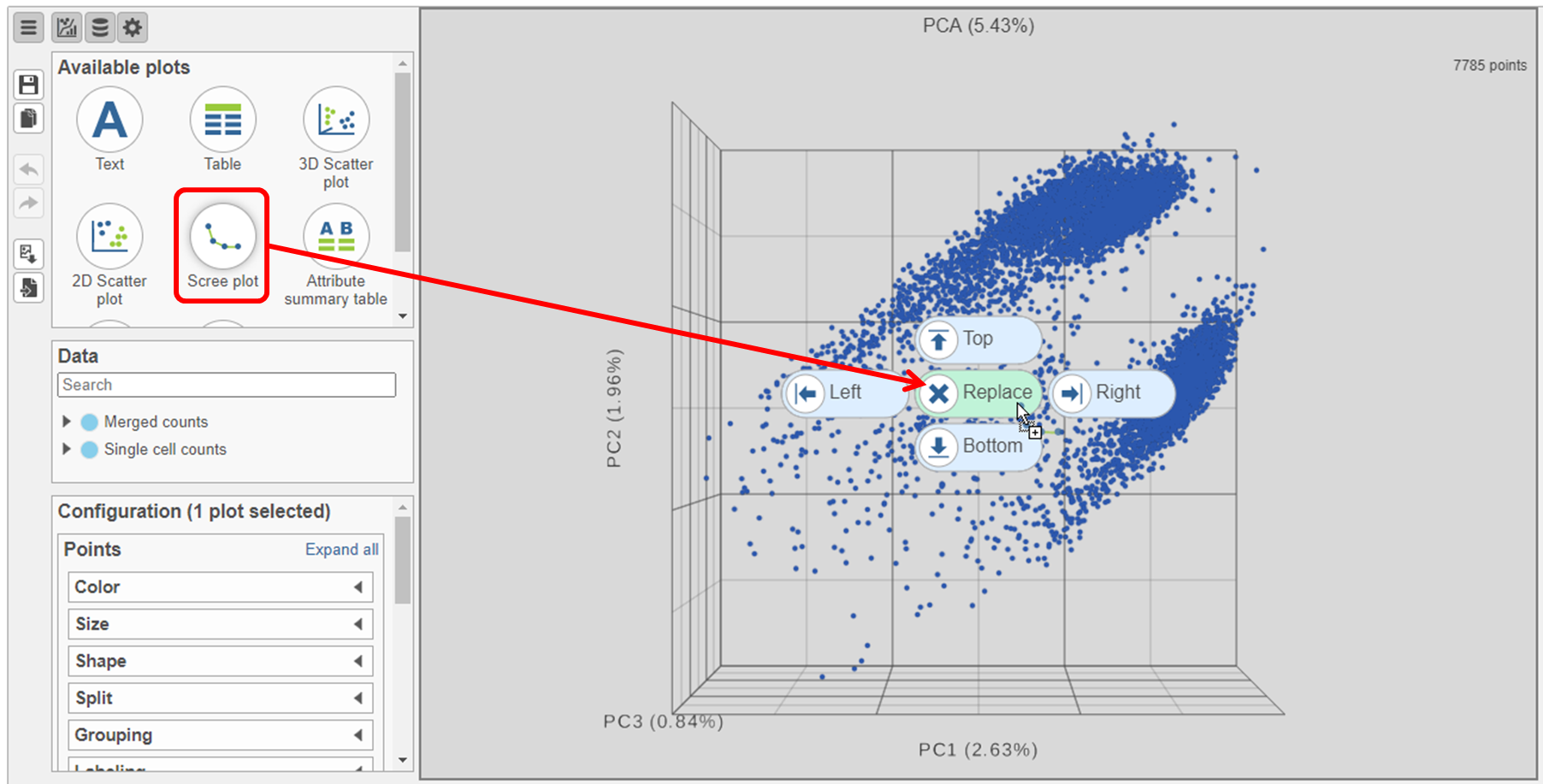 Image Removed Image Removed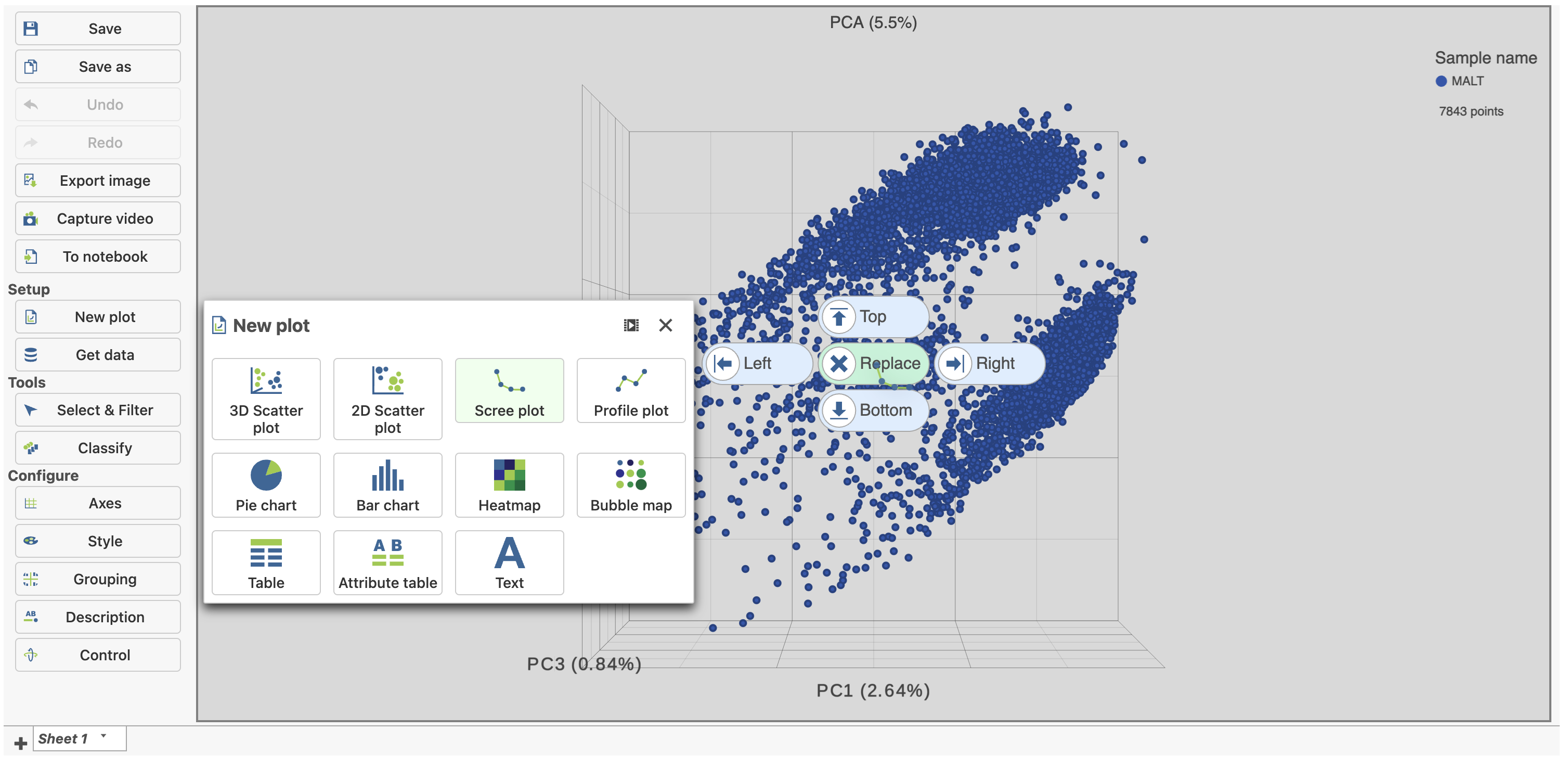 Image Added Image Added
|
- Select PCA as data for the new Scree plot (Figure ?5)
| Numbered figure captions |
|---|
| SubtitleText | The PCA data node contains the data to draw the Scree plot |
|---|
| AnchorName | Choose PCA data for Scree plot |
|---|
|
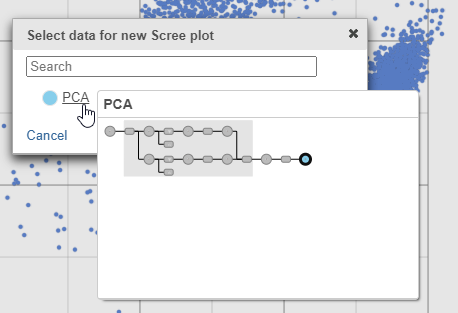 Image Removed Image Removed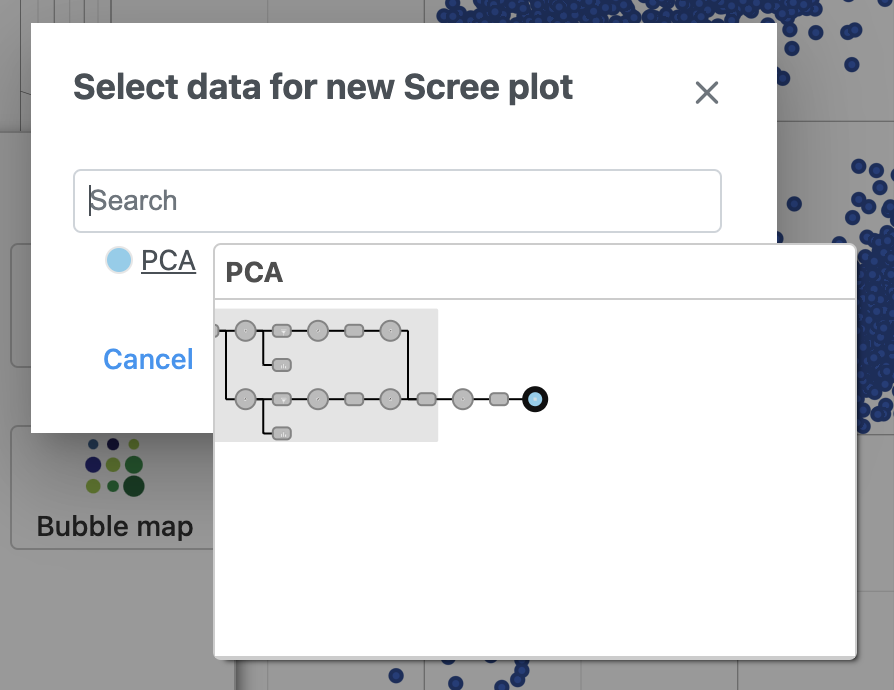 Image Added Image Added
|
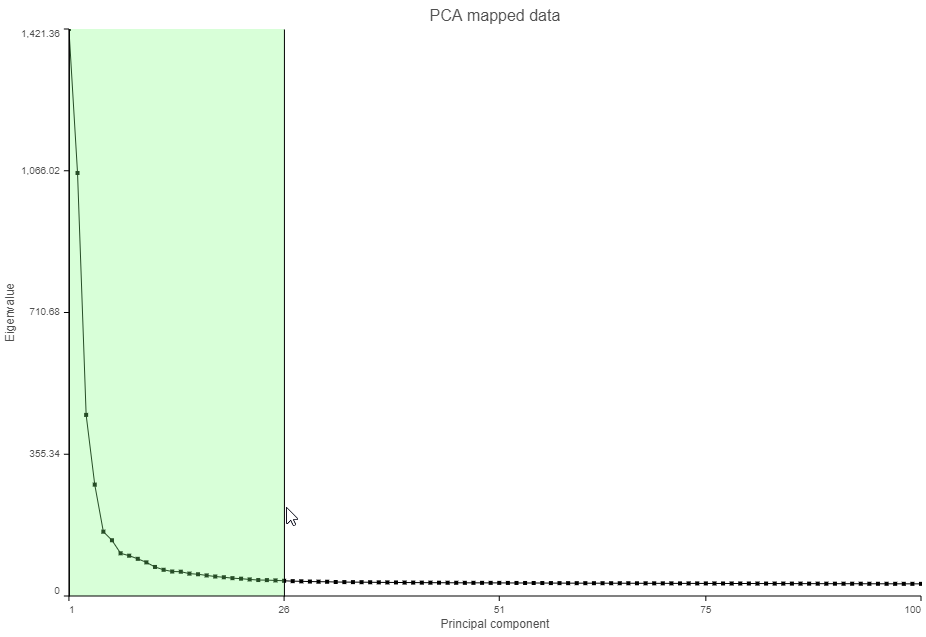
The Scree plot (Figure ?6) shows the eigenvalues on the y-axis for each of the 100 PCs on the x-axis. The higher the eigenvalue, the more variance explained by each PC. Typically, after an initial set of highly informative PCs, the amount of variance explained by analyzing additional components is minimal. By identifying the point where the Scree plot levels off, you can choose an optimal number of PCs to use in downstream analysis steps like graph-based clustering and UMAP.
...
- Click and drag over the first set of PCs to zoom in (Figure ?7)
| Numbered figure captions |
|---|
| SubtitleText | Click and drag on the Scree plot to zoom in and see the first set of principal components |
|---|
| AnchorName | Scree plot zoom in |
|---|
|
|
- Mouse over the Scree plot to identify the point where additional PCs offer little additional information (Figure ?8)
In this data set, a reasonable cut-off could be set anywhere between around 10 and 30 PCs. We will use 15 in downstream steps.
...
| Numbered figure captions |
|---|
| SubtitleText | Identifying the optimal number of PCs |
|---|
| AnchorName | Scree plot PC15 |
|---|
|
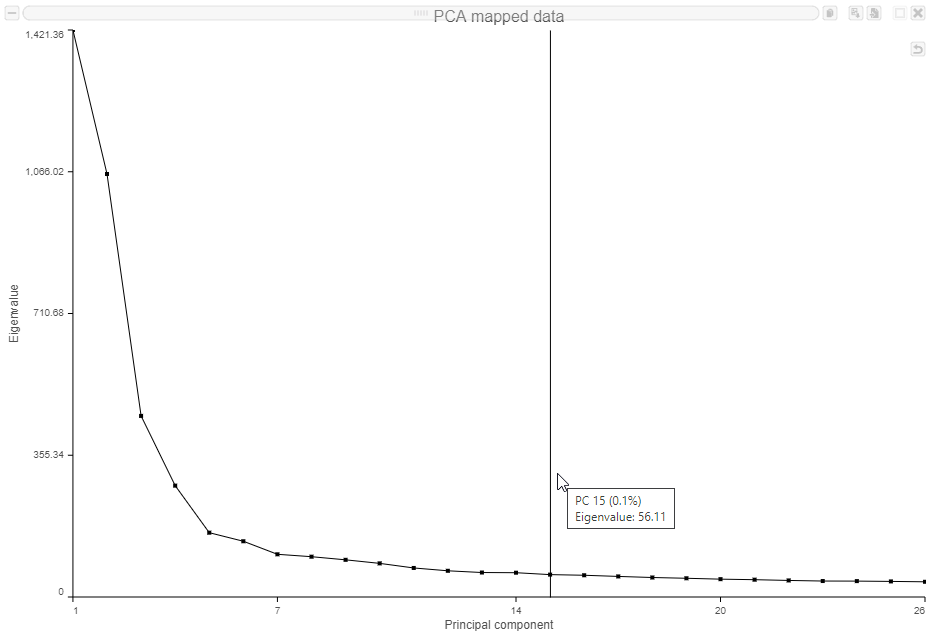 Image Removed Image Removed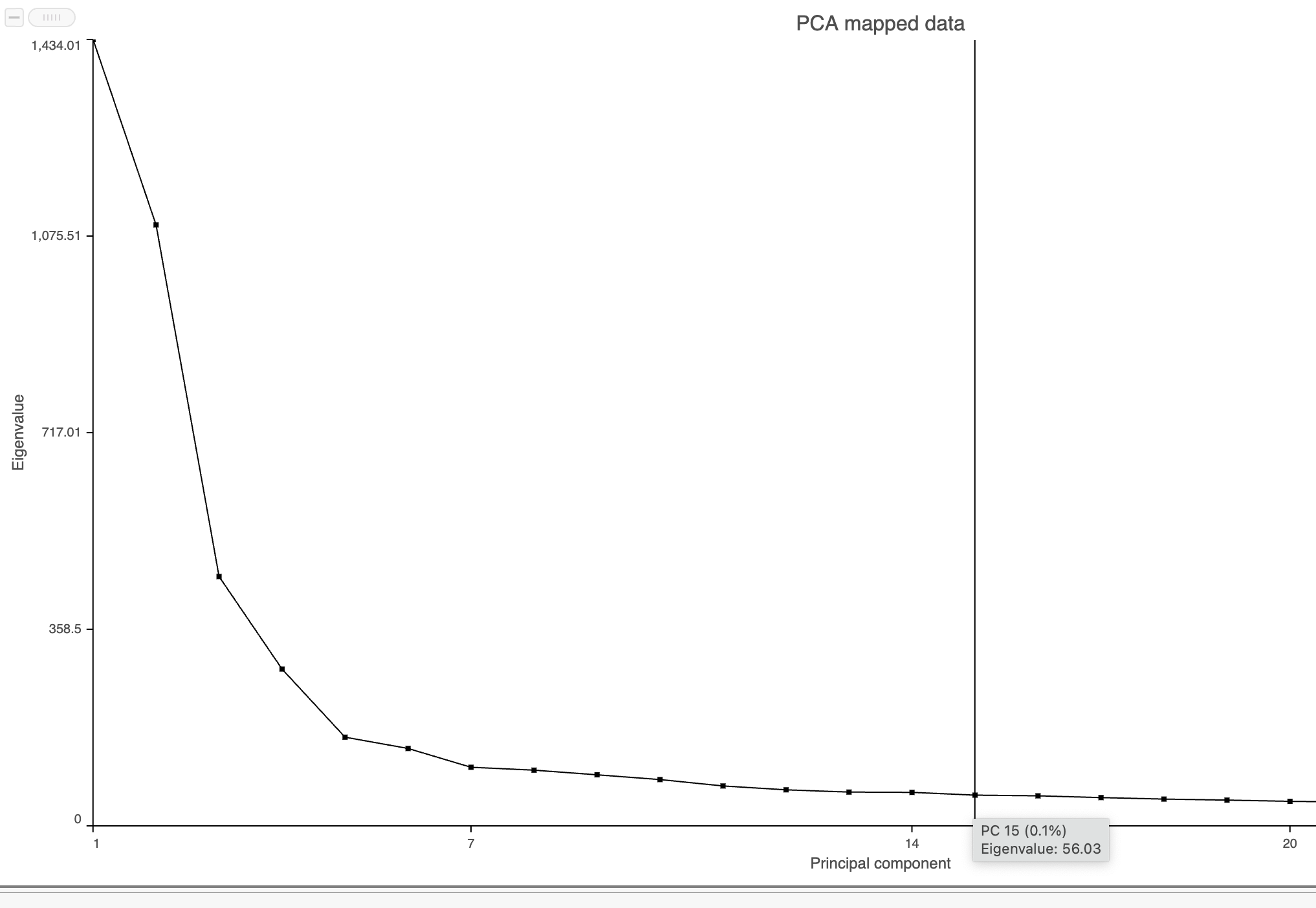 Image Added Image Added
|
Graph-based clustering
...
- Click the project name near the top to go back to the Analyses tab
- Click the circular PCA data node
- Click Exploratory analysis in the toolbox
- Click Graph-based clustering
- Click to Compute biomarkers
- Set the number of principal components to 15 (Figure ?9)
- Click Configure under Advanced options and change the Resolution to 1.0
- Click Finish to run the task
| Numbered figure captions |
|---|
| SubtitleText | Graph-based clustering task set up. Reduce the number of PCs to 15 |
|---|
| AnchorName | Graph-based clustering set up |
|---|
|
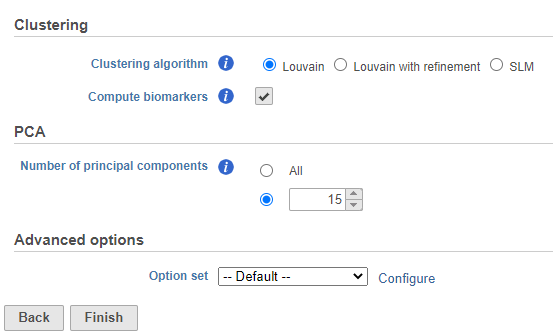 Image Removed Image Removed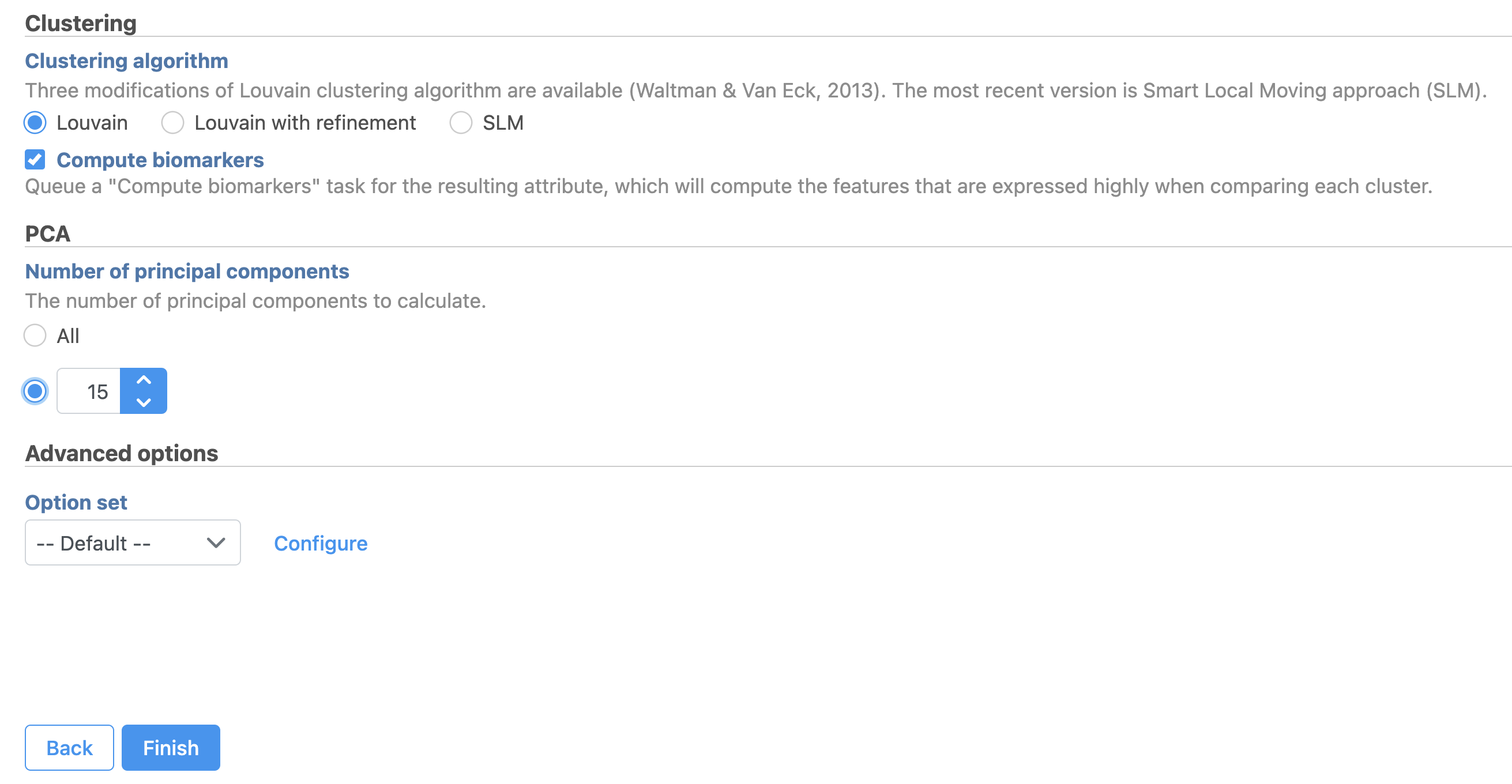 Image Added Image Added
|
A Graph-based clustering task node will be added to the pipeline under the Analyses tab and a circular Graph-based clusters output data node will be produced (Figure ?10)
| Numbered figure captions |
|---|
| SubtitleText | Graph-based clustering task and output data nodes |
|---|
| AnchorName | Graph-based clustering output |
|---|
|
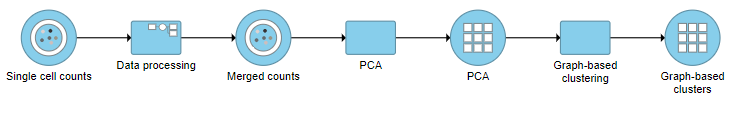 Image Removed Image Removed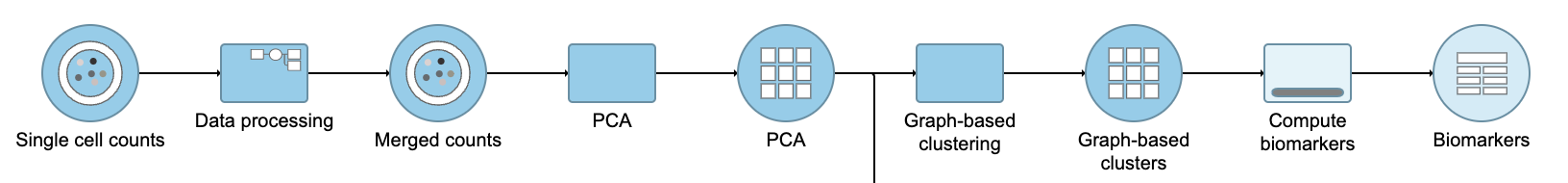 Image Added Image Added
|
UMAP
Once the graph-based clustering task has completed, we can visualize the results with a UMAP plot. You could use the same steps here to generate a t-SNE plot. For this tutorial, we will use UMAP, as it is faster on several thousand cells.
- Click the circular Graph-based clusterscircular PCA data node
- Click Exploratory analysis in the toolbox
- Click UMAP
- Set the number of principal components to 15 (Figure ?11)
- Click Finish to run the task
...
| Numbered figure captions |
|---|
| SubtitleText | UMAP task set up. Reduce the number of PCs to 15. |
|---|
| AnchorName | UMAP task set up |
|---|
|
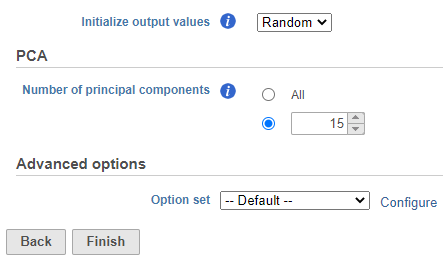 Image Removed Image Removed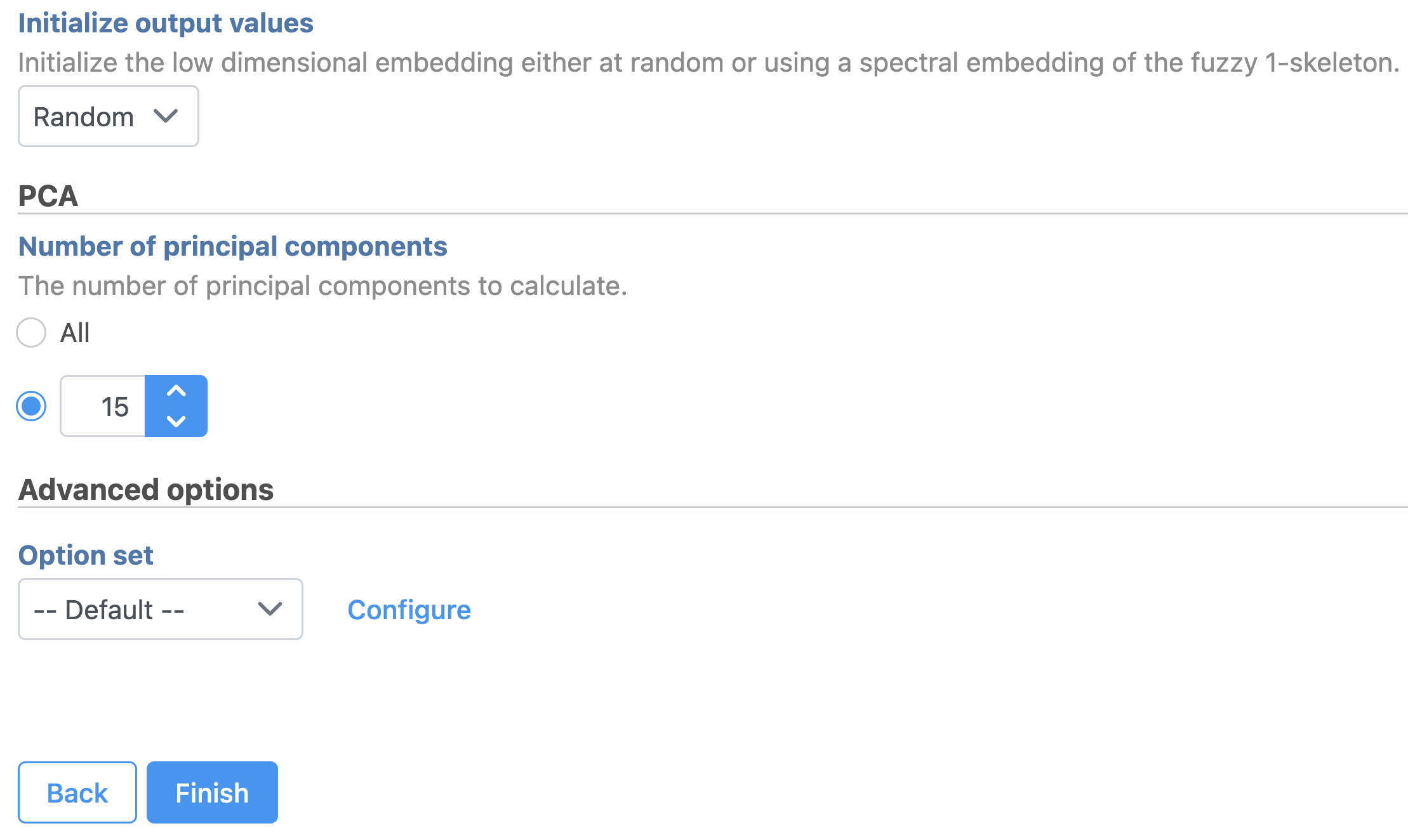 Image Added Image Added
|
A UMAP task node will be added to the pipeline under the Analyses tab and a circular UMAP output data node will be produced (Figure ?12)
| Numbered figure captions |
|---|
| SubtitleText | UMAP task and output data node |
|---|
| AnchorName | UMAP output |
|---|
|
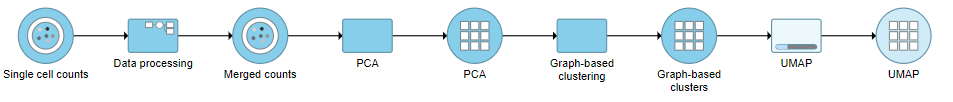 Image Removed Image Removed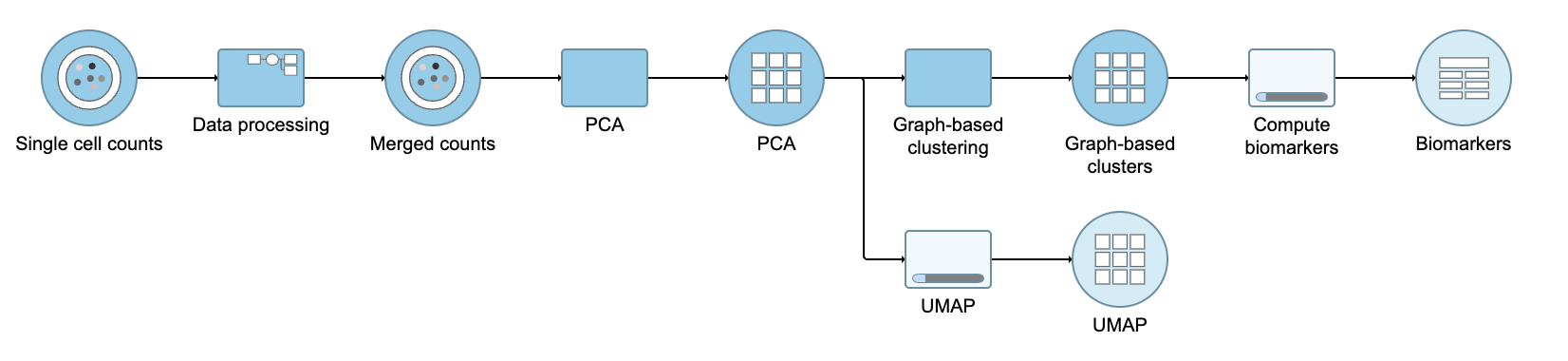 Image Added Image Added
|
Notes on Performing Exploratory Analysis with Protein or Gene Expression Data Only
...
It can be interesting to perform exploratory analysis on the two feature types separately. For example, you might be interested to see how the clustering of the same cells differs between protein expression profiles vs. gene expression profiles. To do this
To perform exploratory analysis on the two feature types separately, select the Merged counts data node, click Pre-analysis tools, followed by Split by feature type from the toolbox. A new task, Split by feature type, will be added to the pipeline resulting in two output data nodes: Antibody capture (protein data) and Gene expression (mRNA data). Both contain the same high-quality cells.
Performing exploratory analysis with gene expression data is the same as for the merged counts. Because there are a large number of genes, you will need to reduce the dimensionality with PCA, choose an optimal number of PCs and perform downstream clustering and visualization (e.g. graph-based clustering and UMAP/t-SNE). Performing exploratory analysis with protein data is different. There is no need to reduce the dimensionality as there are only a handful of features (17 proteins in this case), so you can proceed straight to downstream clustering and visualization. Figure ? 13 shows an example of how the pipeline might look if the data is split and analyzed separately.
...
| Numbered figure captions |
|---|
| SubtitleText | Example of how the pipeline might look if you split the merged counts and perform exploratory analysis for protein and gene expression data separately |
|---|
| AnchorName | Split merged counts for exploratory analysis |
|---|
|
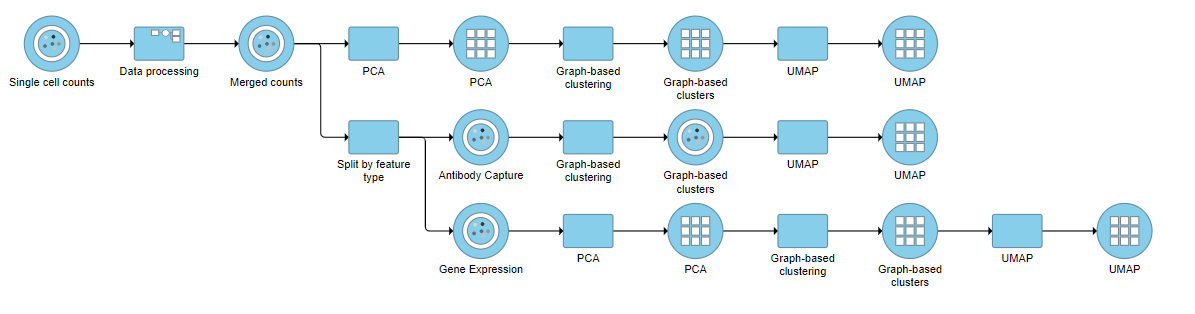 Image Removed Image Removed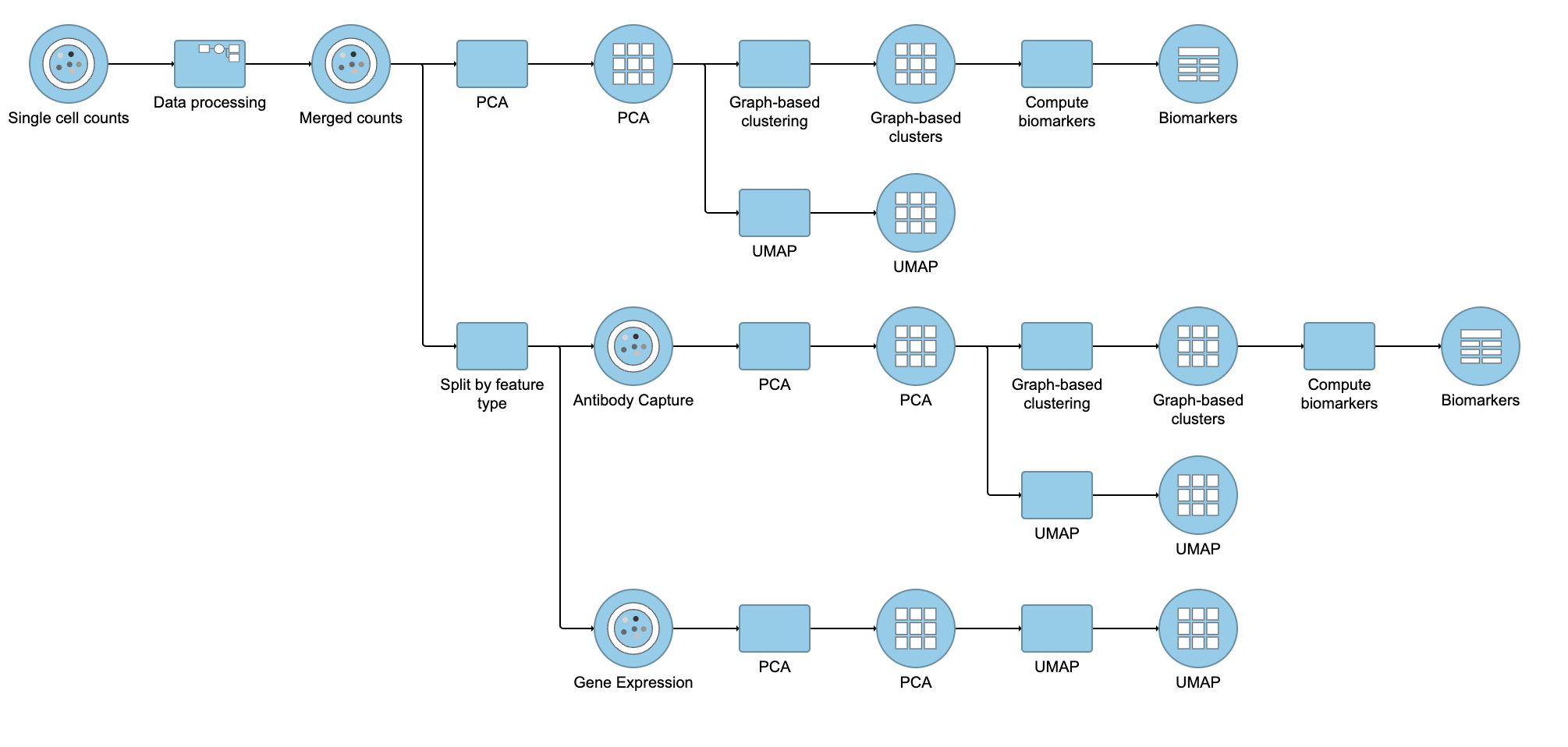 Image Added Image Added
|
You can then use the Data viewer to bring together multiple plots for comparison (Figure ?14).
| Numbered figure captions |
|---|
| SubtitleText | Comparison of 2D UMAP plots for the same cells clustered on protein, mRNA and merged data. All cells are coloured based on their expression of the CD3D gene (in blue). Note, the plots in this figure may differ from the default UMAP plots because these are 2D plots. Default UMAP plots re in 3D. |
|---|
| AnchorName | UMAP plot comparison |
|---|
|
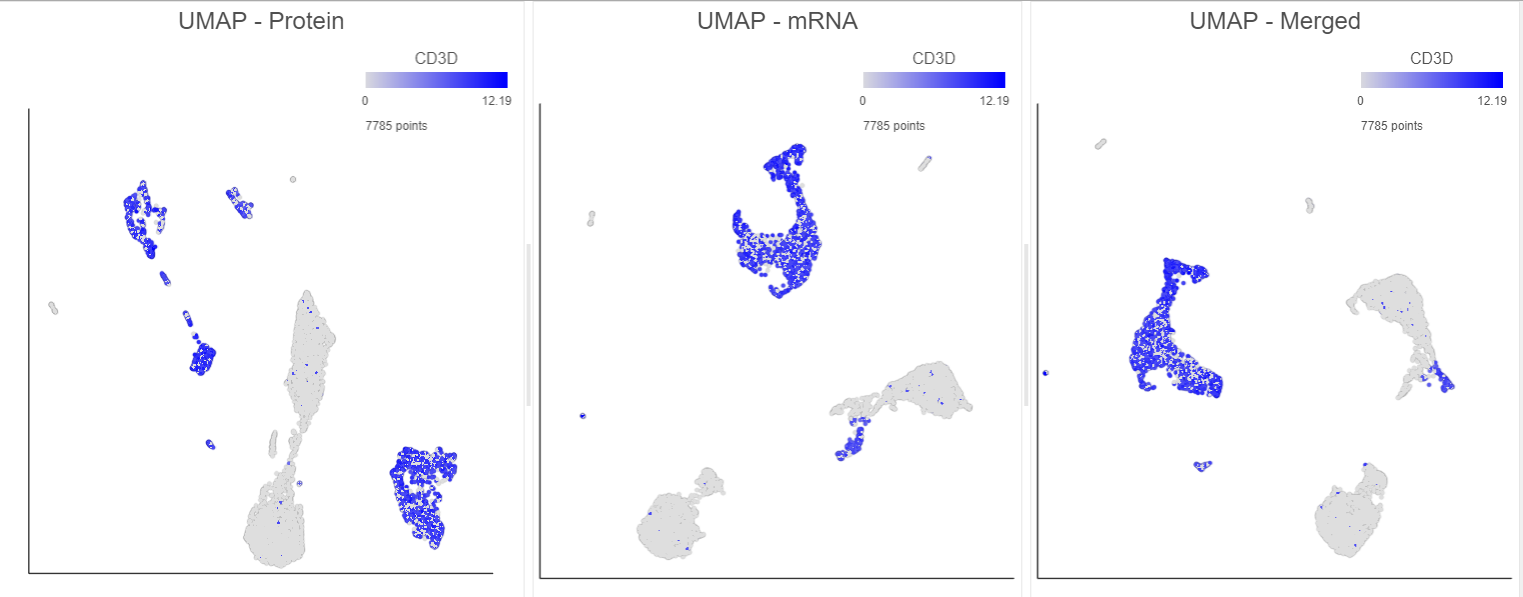 Image Modified Image Modified
|
...





















