Page History
...
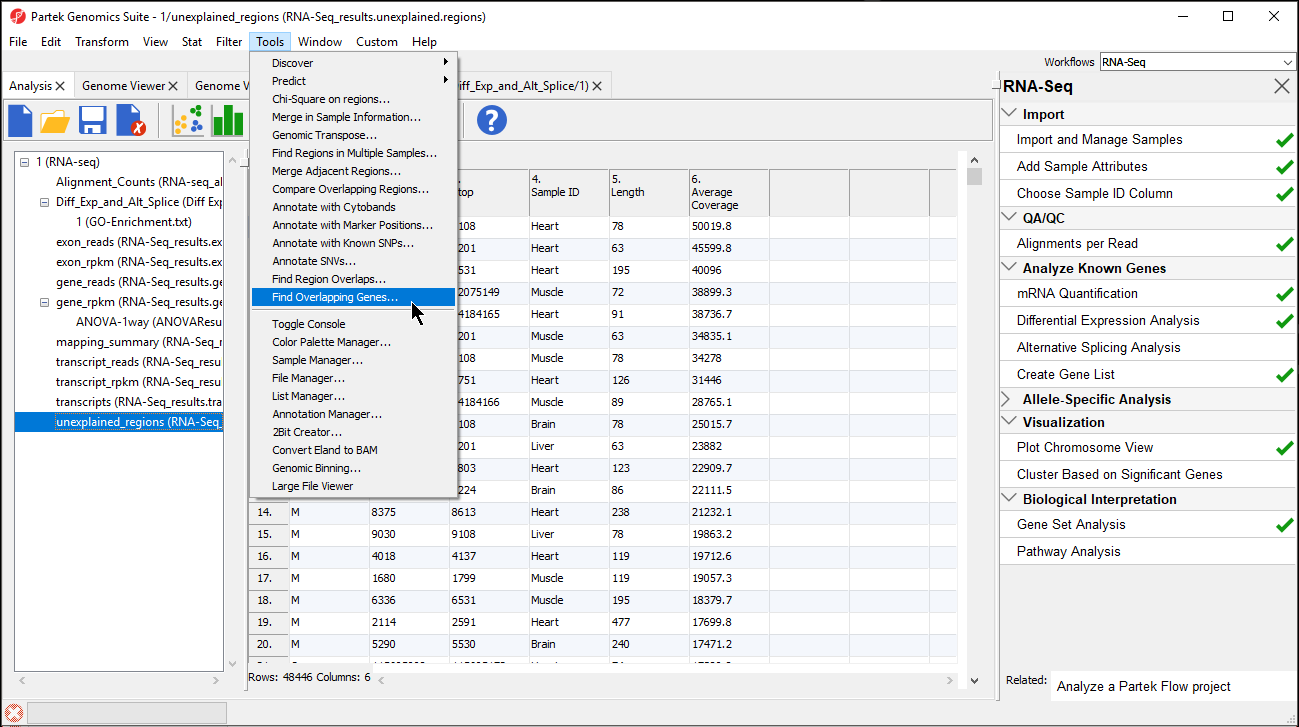
During a previous section of this tutorial, a spreadsheet named unexplained_regions was generated. This spreadsheet contains locations where reads map to the genome but are not annotated by the transcript database, in this case, RefSeqGene. TheunexplainedThe unexplained_regions spreadsheet regions spreadsheet is potentially very interesting as it may contain novel findings.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
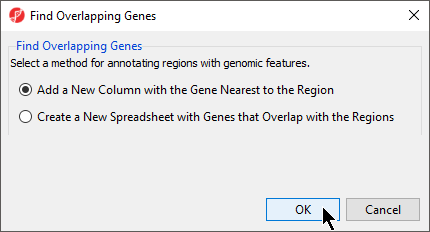
- Select Add a new column with the gene nearest to the region in the Find Overlapping Genes dialog (Figure 2)
- Select OKOK
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
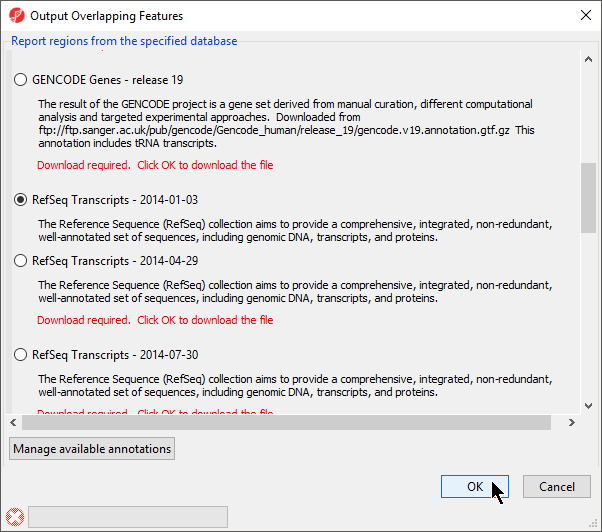
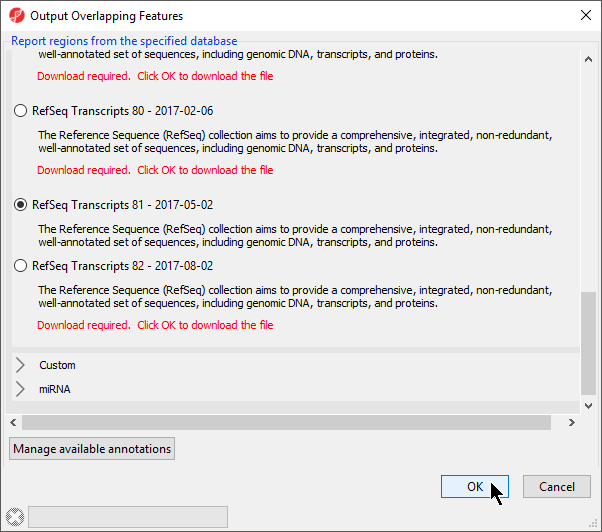
- Select RefSeqTranscripts – 20142017-0105-03 02 from the Output Overlapping Features dialog (Figure 3)
Please note that it is recommended that you annotate with the same database as used when you performed mRNA quantification.
- Select OKOK
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
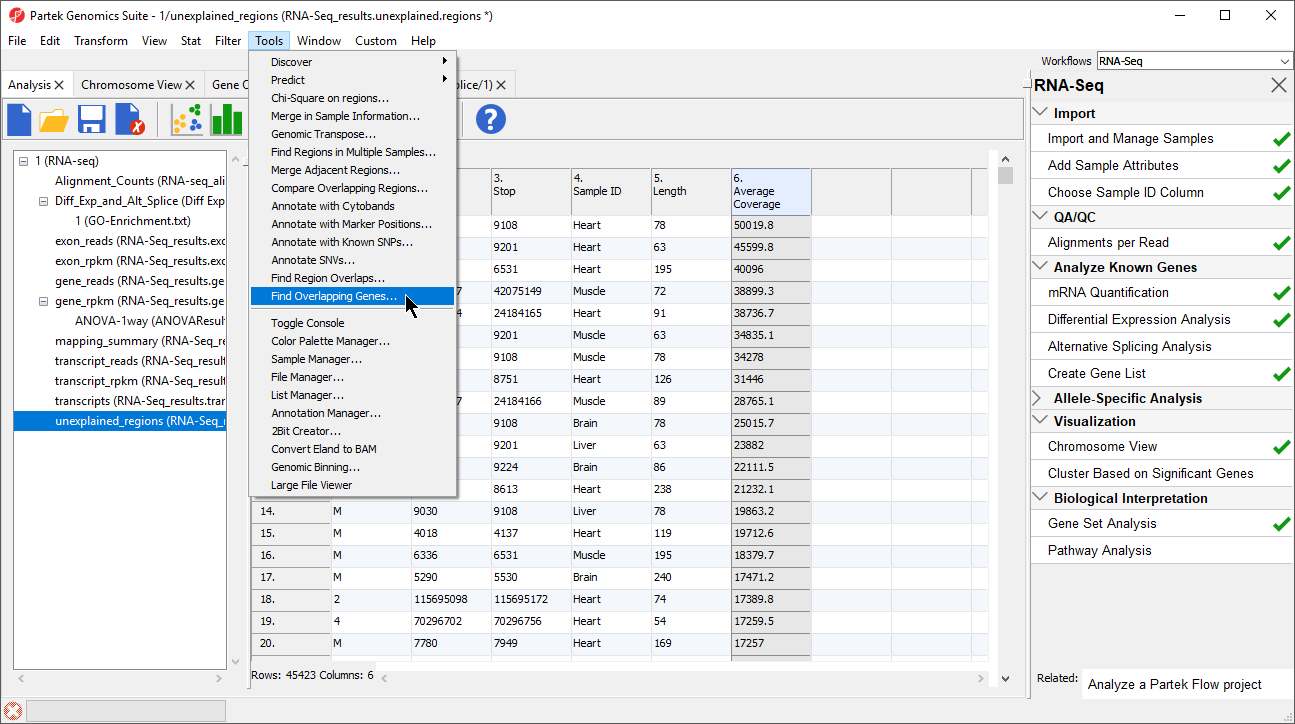
The closest overlapping feature and the distance to it is now included as columns 7. Overlapping Features and 8. Nearest Features in the unexplained_regions spreadsheet.
Right-clicking on a row header and selecting Browse to Location will show the reads mapped to the chromosome. For this tutorial, a couple of genes are selected to show regions that are located after a known gene or in the intron of a gene.
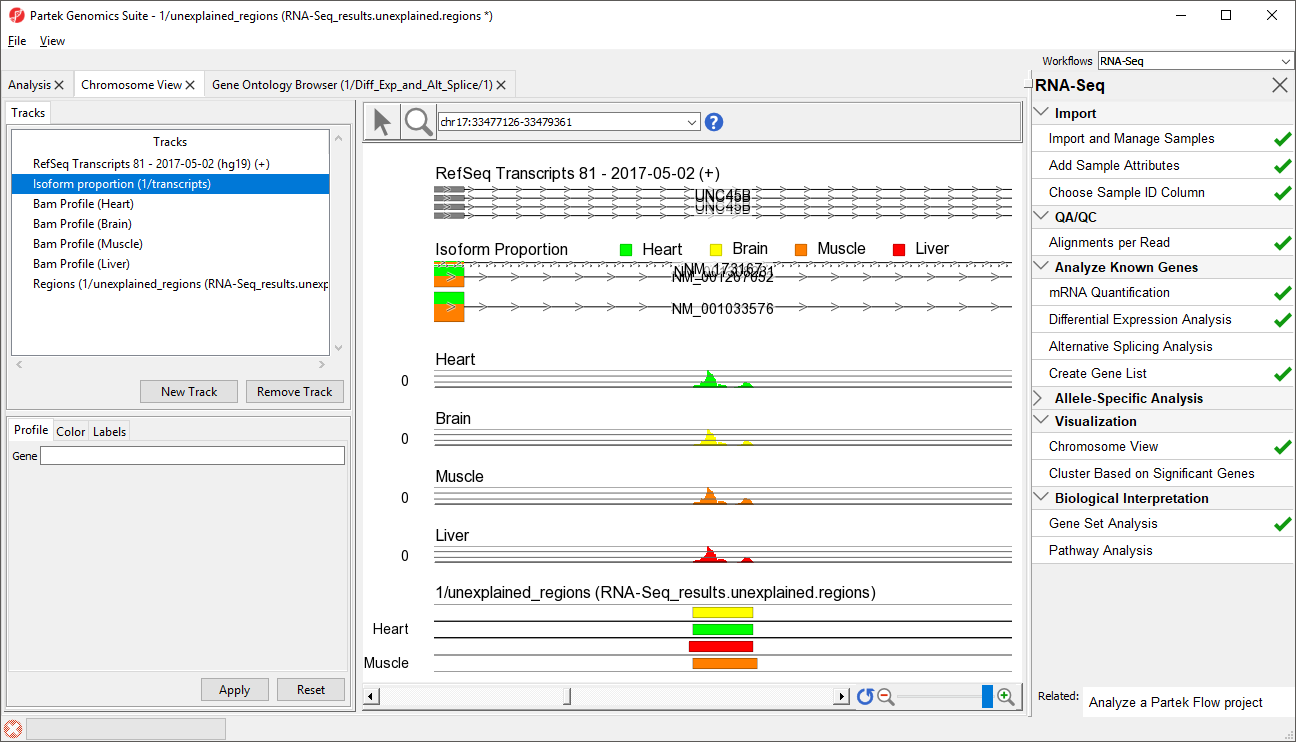
- Right-click row 39 and select Browse to location from the pop-up menu
- Select the Chromosome View tab to view a region within an intron of UNC45B. This may be a novel exon (Figure 4)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
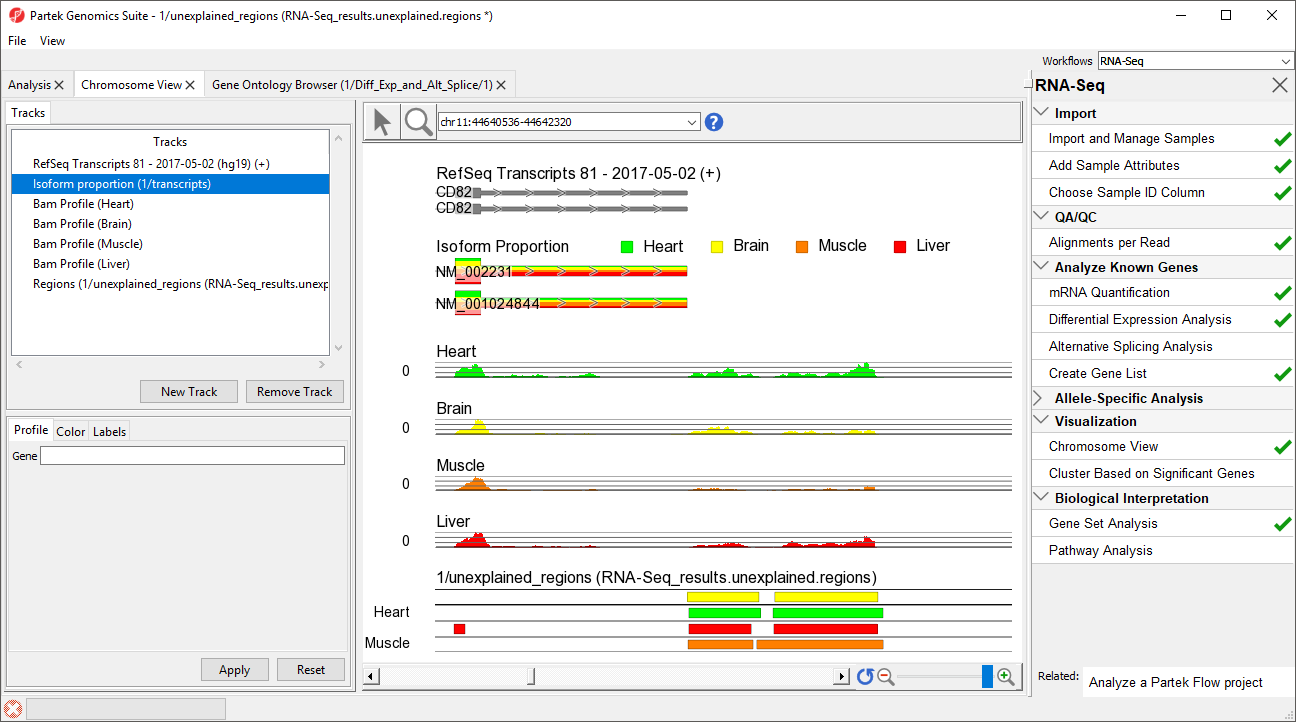
- Right-click row 12576 and select Browse to location to go to a region that starts 1 bp after CD82.
- Select () several times to zoom out slightly
This peak may represent an extended exon (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
While RefSeq was used to identify overlapping features, the choice of which database to use will depend on the biological context of your experiment. For example, you may wish to utilize promoter or miRNA databases if you are interested in regulation of expression.
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|