Page History
...
Count feature barcodes is a tool for quantifying the number of feature barcodes per cell from CITE-Seq, cell hashing, or other feature barcoding assays to measure protein expression. The input for Count feature barcodes is FASTQ files.
...
Count feature barcodes will run on any FASTQ file inputunaligned reads data node.
- Click the data node containing your FASTQ filesunaligned reads containing feature barcodes
- Click the Quantification section in the toolbox
- Click Count feature barcodes
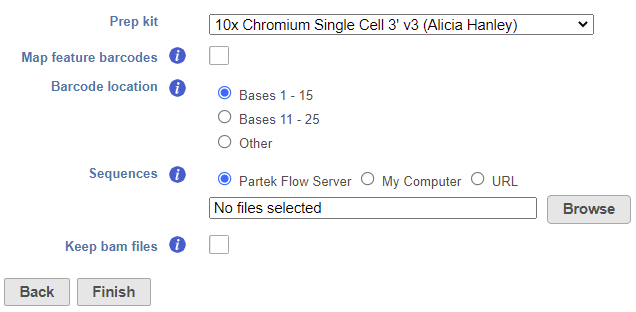
The task set up page allows you to configure the settings for your assay (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Choose the Prep kit from the drop-down menu
For more details on adding Prep kits, please see our documentation on Trim tags. The prep kit should include cell barcode and unique molecular identifiers (UMIs) locations.
- Check Map feature barcodes box (optional)
This is only necessary for processing data from 10X Genomics' Feature Barcoding assay (v3+ chemistry), which utilizes BioLegend TotalSeq-B. For all other assays, leave this box unchecked.
- Choose the Barcode location
...
This tab-delimited text file should have the feature ID in the first column and the nucleotide sequence in the second column. Do not include column headers. See Figure 2 for an example.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Check Keep bam files box (optional)
This option will retain the alignment BAM files instead of automatically deleting them when the task is complete. The BAM files can be found in the Output files section of the Task details. This option is unchecked by default to save on disk space.
- Click Finish to run
The output of Count feature barcodes is a Single cell counts data node.
...
Count feature barcodes uses a series of tasks available independently in Partek Flow to process the input FASTQ files. The output files generated by these tasks are not retained in the Count feature barcodes output. , with the exception of BAM files if Keep bam files is checked.
Trim tags identifies the UMI and cell barcode sequences. The Prep kit is specified using the Prep kit setting.
...