Page History
...
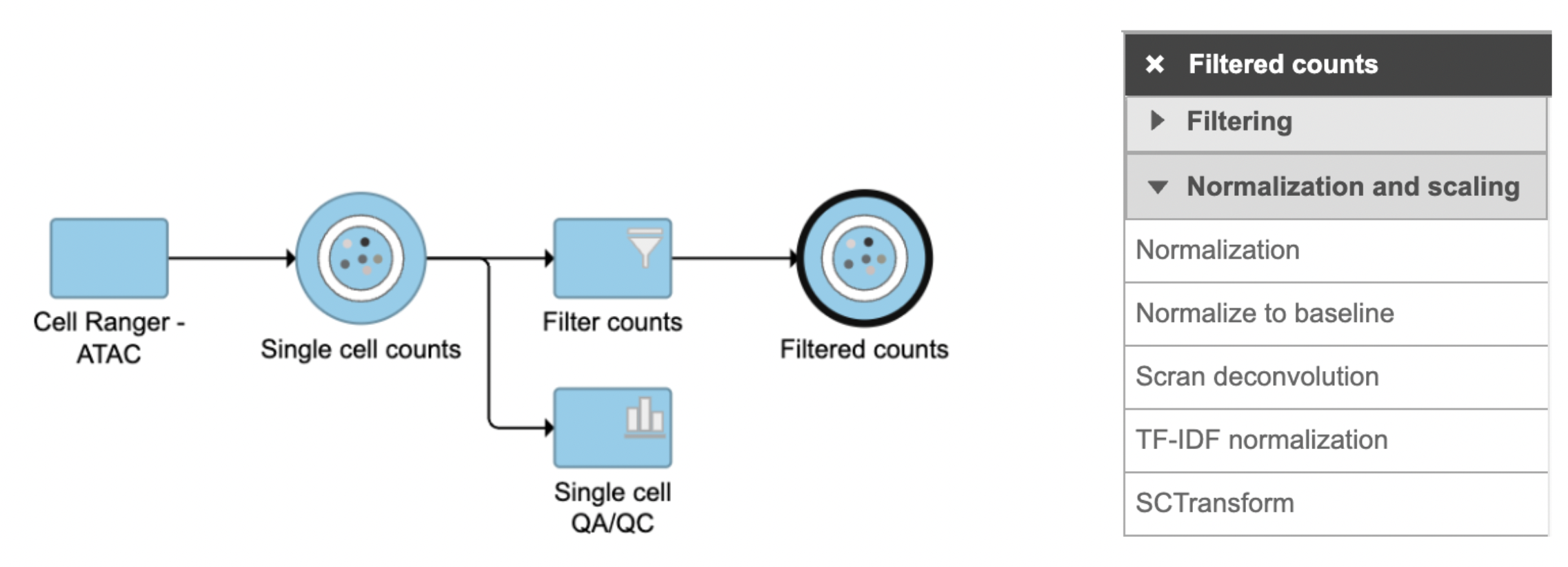
TF-IDF normalization in Flow can be invoked in Normalization and scaling section by clicking any single cell counts data node (Figure 1).
Figure 1. TF-IDF normalization task in Normalization and scaling section in Flow.
...
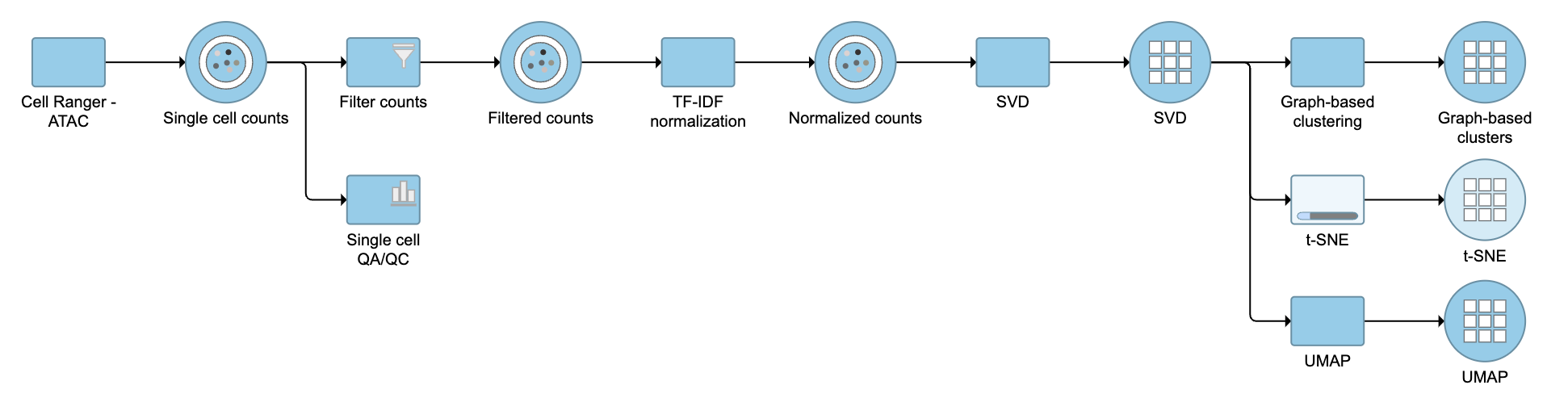
The output of TF-IDF normalization is a new data node that has been normalized by log(TF x IDF). We can then use this new normalized matrix for downstream analysis and visualization (Figure 2).
Figure 32. Example workflows to demonstrate downstream analysis and visualization of Scran deconvolution output.
Other parameters in this task that you can adjust include:
Pool size: A numeric vector of pool sizes, i.e., number of cells per pool.
Max cluster size: An integer scalar specifying the maximum number of cells in each cluster.
Enforce positive estimates: A logical scalar indicating whether linear inverse models should be used to enforce positive estimates.
Scaling factor: A numeric scalar containing scaling factors to adjust the counts prior to computing size factors.
References
- Lun, A. T., K. Bach, and J. C. Marioni. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016.
...
TF-IDF normalization output.
References
- Cusanovich, D., Reddington, J., Garfield, D. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018). https://doi.org/10.1038/nature25981
- https://satijalab.org/signac/index.html
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.