Page History
...
- To invoke the ANOVA dialog, select Detect Differentially Expressed Genes in the Analysis section of the Gene Expression workflow
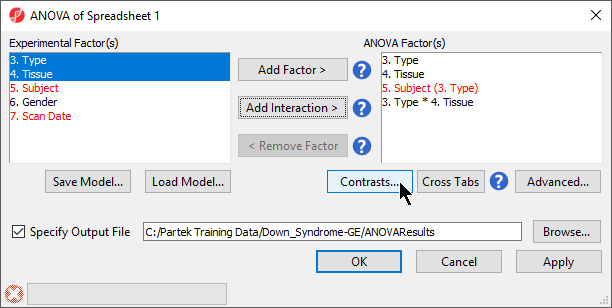
- In the Experimental Factor(s) panel, select Type, Tissue and Subject by pressing <Ctrl> and left clicking each factor
- Use the Add Factor > button to move the selections to the ANOVA Factor(s) panelTo specify
- the interaction, select Select both Type and Tissue by pressing holding <Ctrl> on your keyboard and left clicking each factor.
- Select the Add Interaction > button to add the a Type * Tissue interaction to the ANOVA Factor(s) panel (Figure 1).
Do NOT select OK or Apply. We will be adding contrasts to this ANOVA model in an upcoming section of the tutorial.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Random vs. fixed effects – mixed model ANOVA
...
- Select Contrasts… to invoke the Configure dialog
- Choose 6.Type from the Select Factor/Interaction drop-down list. The levels in this factor are listed on the Candidate Level(s) panel on the left side of the dialog
- Left click to select Down Syndrome from the Candidate Level(s) panel and move it to the Group 1 panel (renamed Down Syndrome) by selecting Add Contrast Level > in the top half of the dialog.
Label 1 will be changed to the subgroup name automatically, but you can also manually specify the label
...
name.
- Select Normal from the Candidate Level(s) panel and move it to the Group 2 panel (renamed Normal)
- The Add Contrast button can now be selected (Figure 2)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
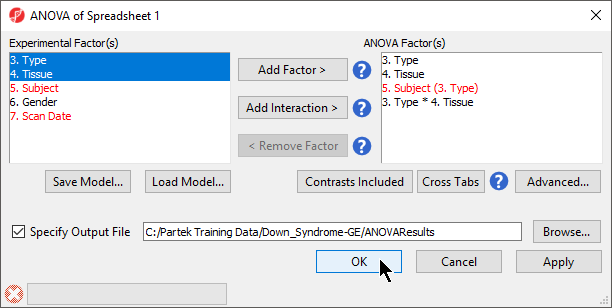
- By default, Specify Output File is checked and gives a name to the output file. If you are trying to determine which factors should be included in the model and you do not wish to save the output file, simply uncheck this box
- Select OK in the ANOVA dialog to compute the 3-way mixed-model ANOVA
Several progress messages will display in the lower left-hand side of the ANOVA dialog while the results are being calculated.
The result will be displayed in a child spreadsheet, ANOVA-3way (ANOVAResults). In this spreadsheet, each row represents a probe set and the columns represent the computation results for that probe set (Figure 4). Although not synonymous, probe set and gene will be treated as synonyms in this tutorial for convenience. By default, the genes are sorted in ascending order by the p-value of the first categorical factor. In this tutorial,Type is the first categorical factor, which means the most highly significant differently expressed gene between Down syndrome and normal samples is at the top of the spreadsheet in row 1.
...
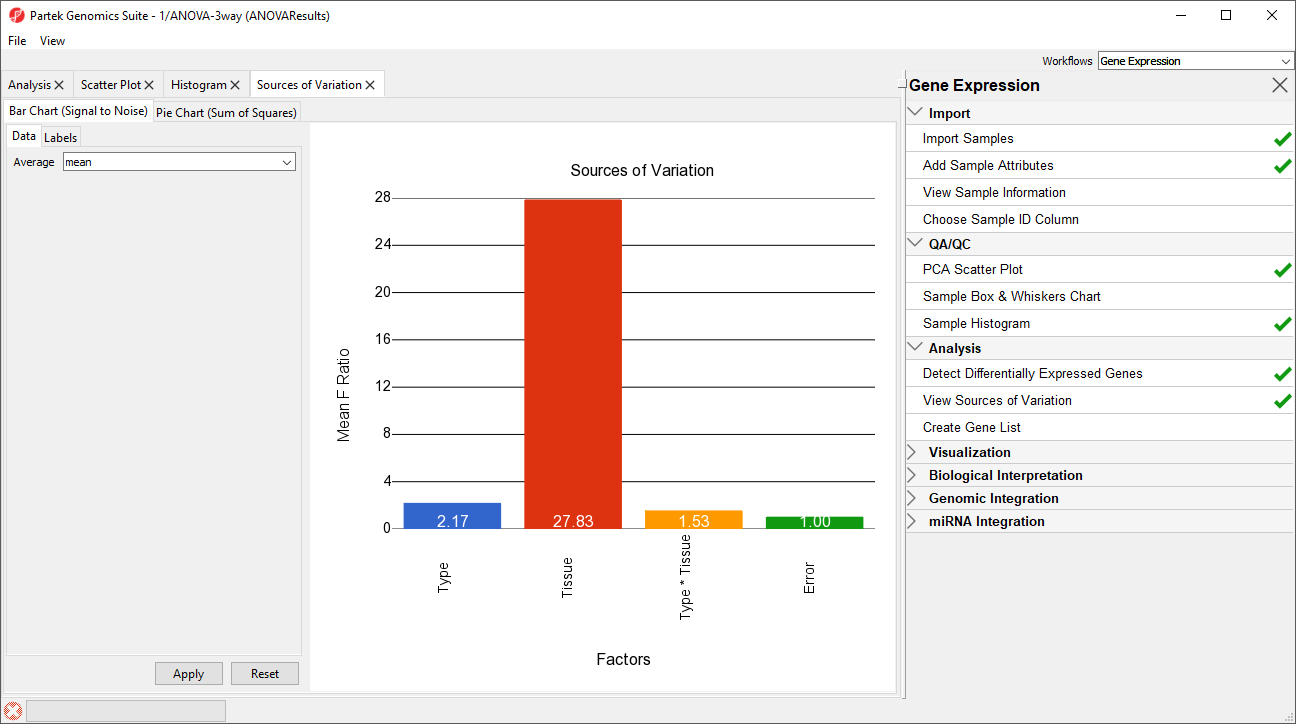
Deciding which factors to include in the ANOVA may be an iterative process while you decide which factors and interactions are relevant as not all factors have to be included in the model. For example, in this tutorial, Gender and Scan date were not included. The Sources of Variation plot is a way to quantify the relative contribution of each factor in the model towards explaining the variability of the data.
- View the sources of variation for each of the factors across the whole genome by clicking Select Plot Sources of Variation from the Analysis section of the Gene Expression workflow with the ANOVA result spreadsheet activeactive
A Sources of Variation tab will appear (Figure 5) with a bar chart showing the signal to noise ratio for each factor accross the whole genome. Sources of variation can also be viewed as a pie chart showing sum or squares by selecting the Pie Chart (Sum of Squares) tab in the upper left-hand side of the Sources of Variation tab.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...