Page History
Table of Contents maxLevel 2 minLevel 2 exclude Additional Assistance
A list of SNPs using dbSNP IDs can be imported as a text file and associated with an annotation file as described for a list of genes. The annotation file (genomic database) you use to annotate the SNPs should minimally contain the genomic coordinates ( chromosome number and physical position ) of the each locus.
Novel SNPs, or SNPs that are not found in your annotation source, must be imported as a region list. The only difference would be to use the SNP name in place of a region name.
Annotating SNPs with genes
Starting with a list of SNPs that have been associated with genomic loci using an annotation file and assigned a species with genome build, you may use Find Overlapping Genes to annotate these SNPs with the closest genes.
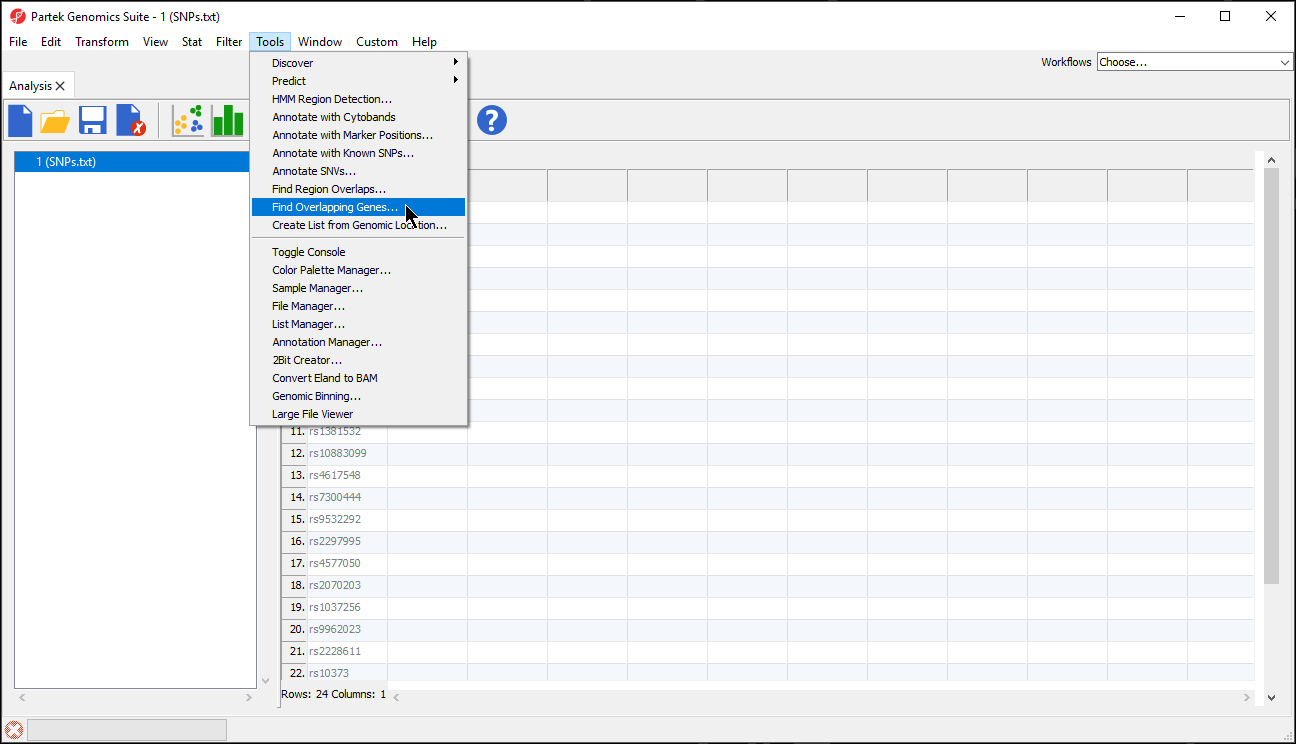
- Select Tools from the main toolbar
- Select Find Overlapping GenesGenes (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Add a New Column with the Gene Nearest to the Region from the method dialogdialog
The Report Regions from the specified database dialog will open.
- Select your preferred database. Be sure to match the species and genome build of your SNP list
- Select OK
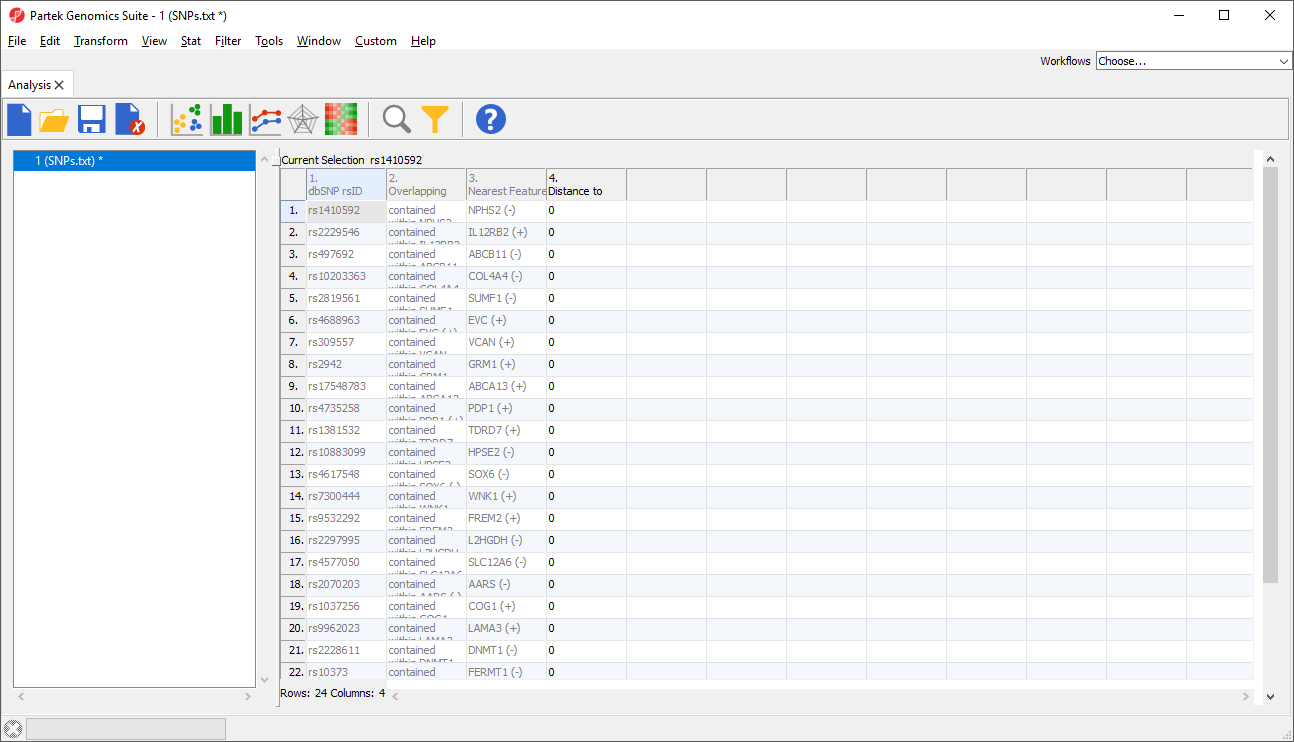
This will add 3 columns to the list of SNPs spreadsheet including Nearest Feature, which will indicate the nearest gene and strand (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To allow gene list operations such as GO Enrichment or Pathway Enrichment to be performed on the SNP list, we can set the Nearest Feature column as the Gene Symbol columnthe gene symbol column for the spreadsheet.
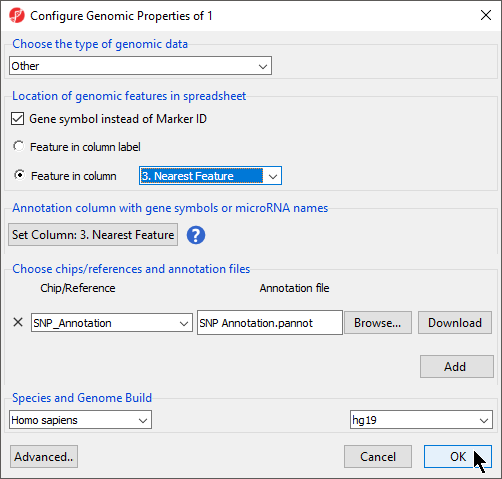
- Right click the spreadsheet in the spreadsheet tree
- Select Properties from the pop-up menu
- Select Gene symbol instead of Marker ID
- Select Feature in column and select Nearest FeatureFeature (Figure 3)
- Select OKOK
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Annotating a Partek Genomics Suite-generated SNP list with SNVs
If you have a SNP spreadsheet that was generated using Partek Genomics Suite (not imported as a .txt file), you can annotate the SNP list with gene, transcript, exon, and information about the predicted effect of the SNPs.
- Select Tools from the main command toolbar
- Select Annotate SNVs
| Page Turner | ||
|---|---|---|
|
...