Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
A BED (Browser Extensible Data) file is a special case of a region list: it is a tab-delimited text file and the first three columns of BED files contain the chromosome, start, and stop locations. To import a bed file to be used as a data region list, follow the import instructions for region lists. A BED File might also be visualized as an annotation file containing regions in the Genome Browser.
Using a BED file as an annotation source for the genome browser
BED files do not contain individual sequences nor do the regions have names. For instanceexample, the UCSC table browser has a BED file that contains reads from a long non-coding RNA-Seq experiment and you Genome Browser has an annotation BED file for CpG islands. You might like to view this information in the context of your dataseta methylation microarray data set. Before you could can visualize a BED file in the chromosome viewer, you would have to must create a Partek annotation file from the BED file.
- From the top command menu, select Tools > Annotation Manager
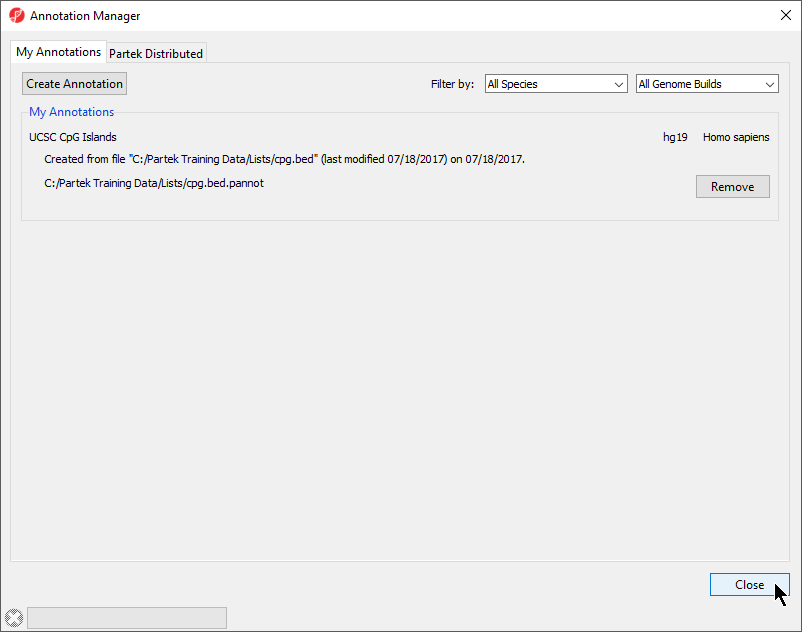
- In the My Annotations tab, select Create Annotation
- Select Tools from the main toolbar
- Select Annotation Manager... (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Create Annotation from the My Annotations tab of the Annotation Manager dialog (Figure 2)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
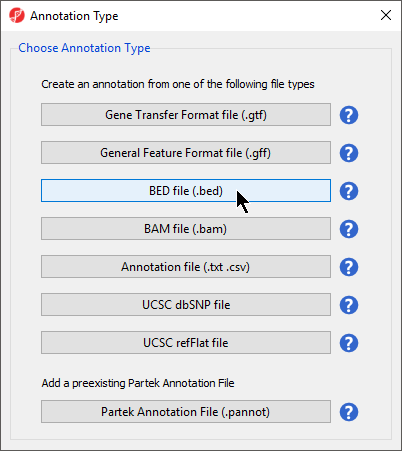
- Select BED file (.bed) under Choose Annotation Type
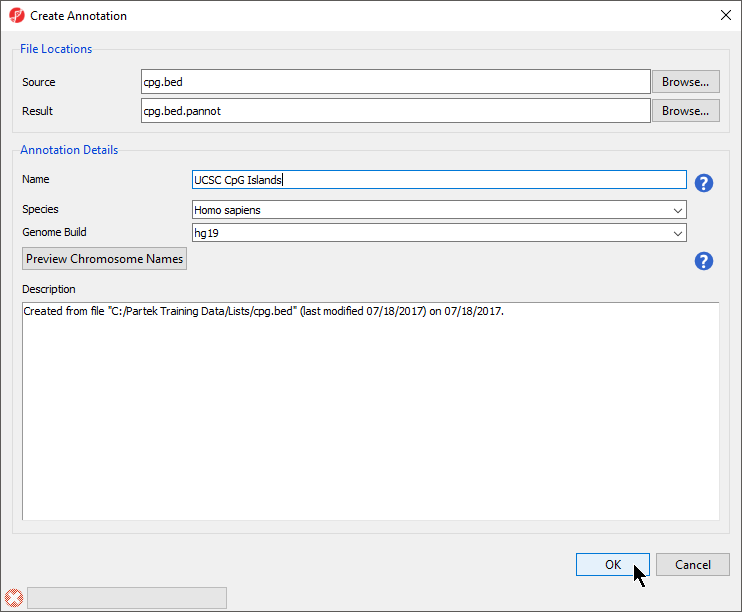
Under File Locations, specify the Source (input BED file) with the Browse menu item. You may also specify the Result file name (of the annotation file) and location with the Browse button. You might consider saving the result file to your Microarray Libraries folder
In the Annotation Details section of the dialog box, specify the Name of the annotation database that will be visible from within Genomics Suite, Species, and Genome Build. Preview Chromosome Names would be used if the chromosome names in the annotation file must be changed to match the name of the chromosome in the genomic annotations.
Select OK
- for Annotation Type (Figure 2)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Browse... under Source to specify the BED file; a default new file name and destination will populate Result, but this can be changed
- You can specify the name and save location of the new annotation file under Result; we typically choose the Microarray Libraries folder
- Specify the Name of the annotation database file
- Select the correct Species and Genome Build for the annotation file from the drop-down menus (Figure 3)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Preview Chromosome Names would be used if the original file had chromosome names that did not match the genome build that had required modification. For our example, this is unnecessary.
- Select OK to create the annotation
The Annotation Manager will display the new annotation in the My Annotations tab (Figure 4)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Visualizing a BED file as an annotation track in the genome browser
In order to use a BED file as an Annotation track in the Genome Browser, first create the annotation file as described above, being careful to have specified specify the correct species and genome build appropriately.
- Invoke the Genome Browser by right-clicking in Right-click a row on any spreadsheet that has genomic features on rows (gene lists, ANOVA results, SNP detection) and select
- Select either Browse to Row or Browse to Location
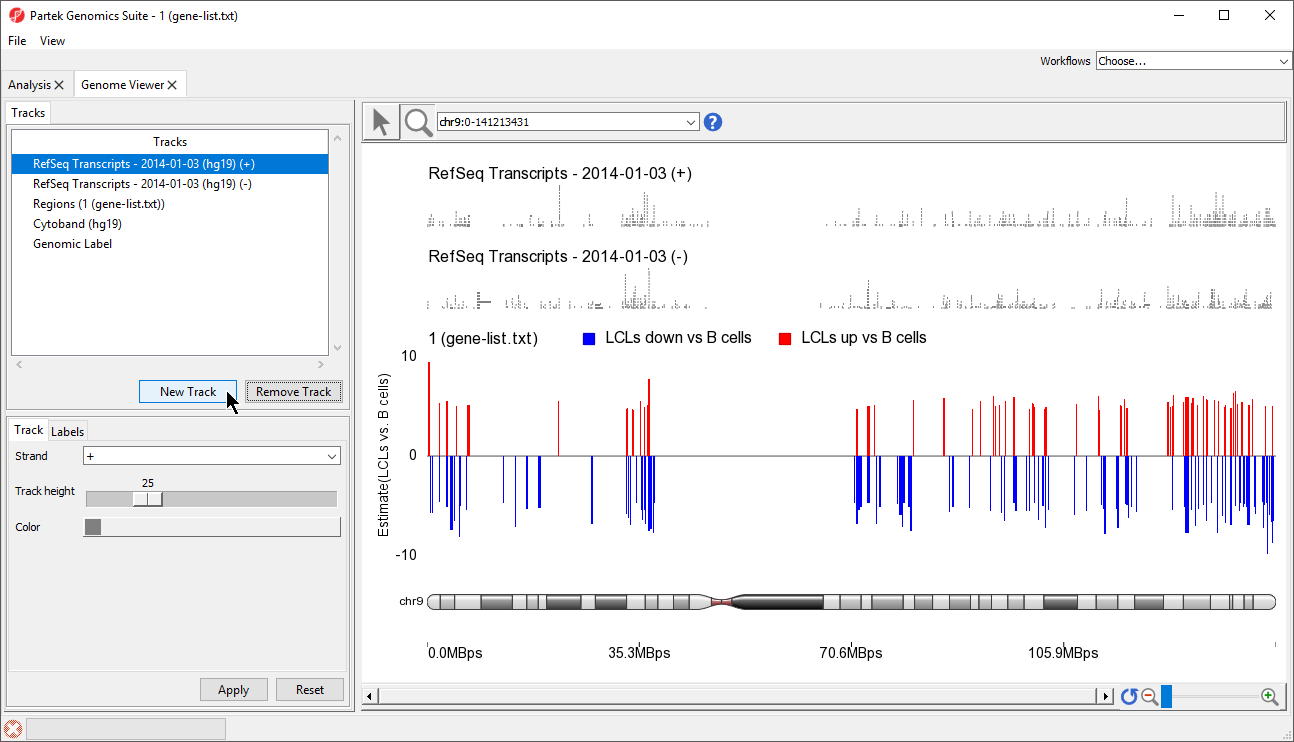
In the Track Toolbar on the left, select New Track which invokes the dialog shown in Figure 21
- Select to invoke the Genome Browser tab
- Select New Track from the Tracks panel of the Genome Browser (Figure 5)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
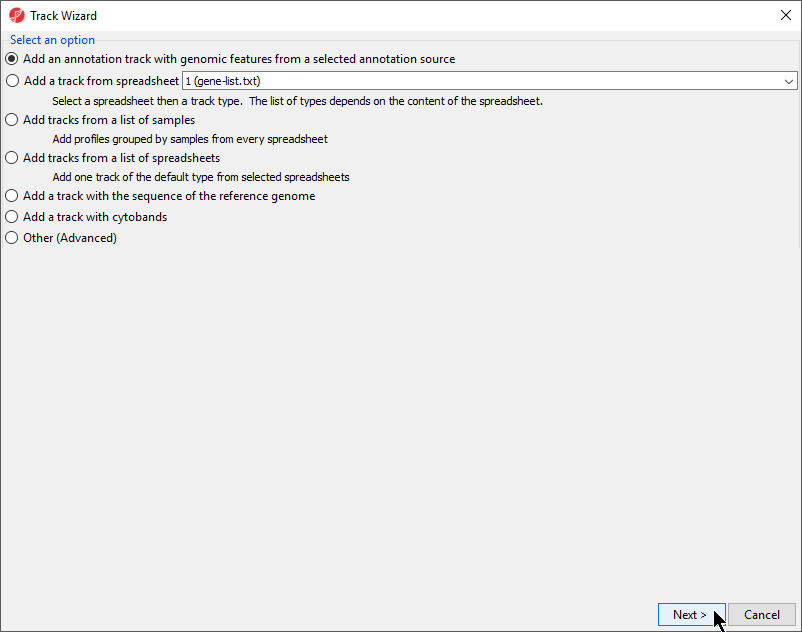
- Select Add an annotation track with genomic features from a selected annotation source and select Next
Next you will have to choose the annotation file that was created from the BED file. The procedure for doing this might vary slightly depending on the type of spreadsheet you have displayed in the Genome Browser. You may be shown a list of annotations that includes the annotation source you have created; in this case, select the radio button in the Available Annotations panel. If, however,
At the bottom of the screen, you should either check or uncheck the box next to Separate strands (checking means that the BED file contained the strand information for each region and you wish to visualize the regions on different strands). Unselecting Separate strands should be used if the BED file did not contain strand information or if you do not wish to display the information on separate strands. Select Create.
- from the Track Wizard dialog (Figure 6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Next >
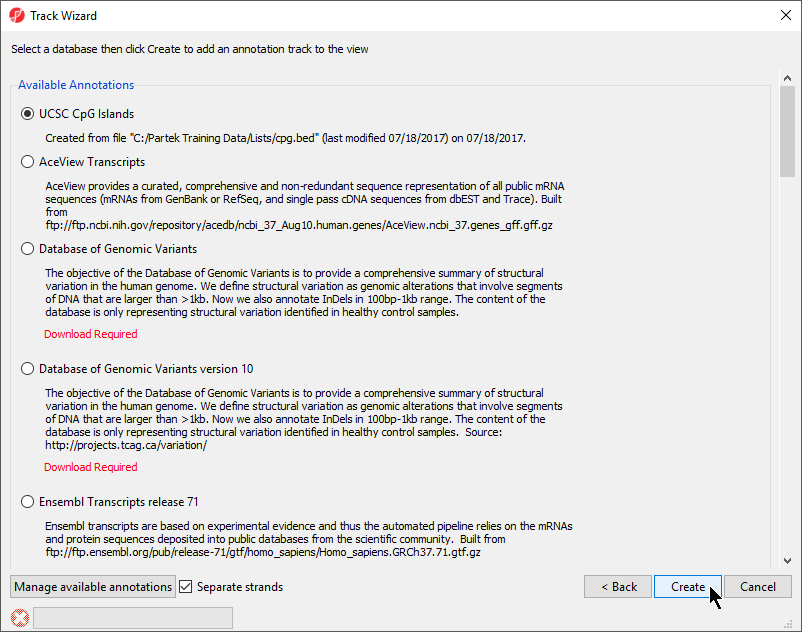
- Choose the annotation file you created; here we have selected UCSC CpG Islands (Figure 7)
- If your annotation file does not contain strand information for each region, deselect Separate Strands; here we have deselected it
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Create
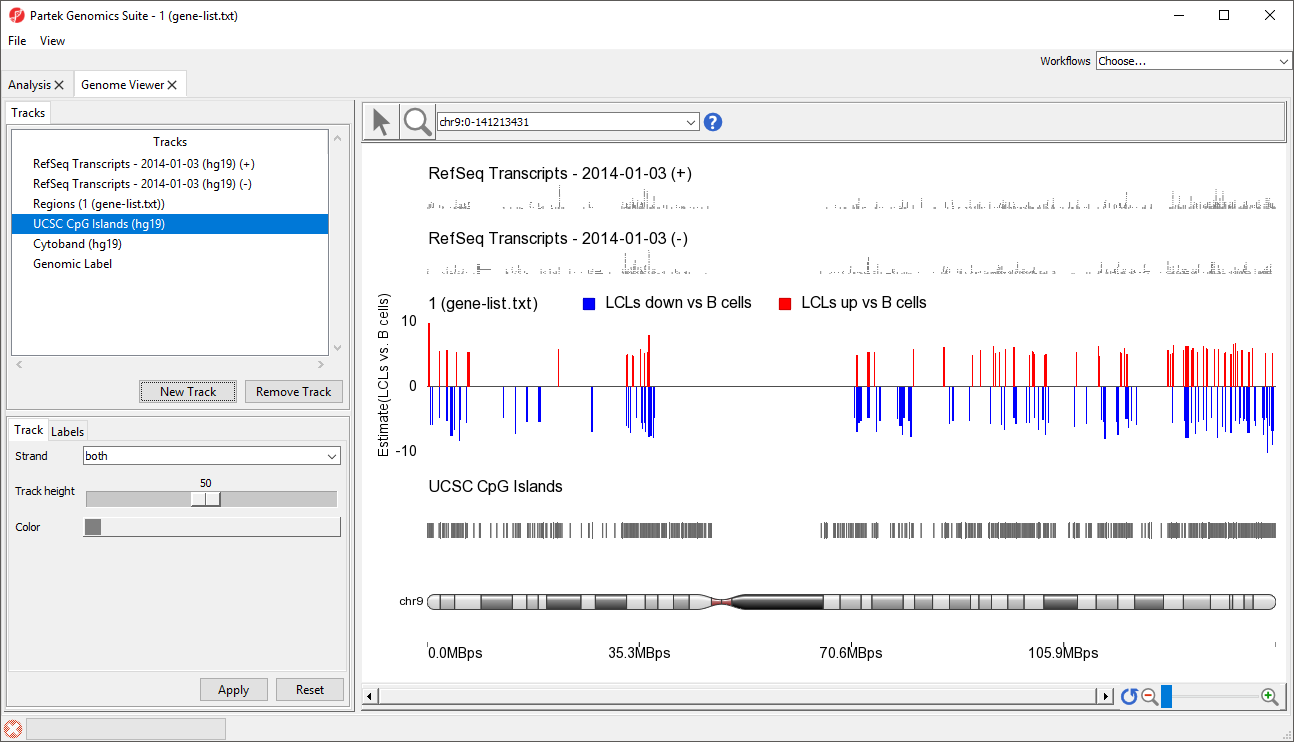
A new track will be created from the annotation file (Figure 8). If Separate Strands had been selected, there would be two tracks, one for each strand, like we see for the RefSeq Transcripts - 2014-01-03 (+) and (-) tracks (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Page Turner | ||
|---|---|---|
|
...