...
| Numbered figure captions |
|---|
| SubtitleText | Select the PCA task from the Exploratory analysis menu |
|---|
| AnchorName | Select PCA task |
|---|
|
 Image Modified Image Modified
|
- Click Finish to run PCA with default settings (Figure 2)
...
| Numbered figure captions |
|---|
| SubtitleText | Invoking t-SNE from the task menu |
|---|
| AnchorName | Invoking t-SNE |
|---|
|
 Image Modified Image Modified
|
- Click Finish from the t-SNE dialog to run t-SNE with the default settings (Figure 4)
...
| Numbered figure captions |
|---|
| SubtitleText | t-SNE task node |
|---|
| AnchorName | t-SNE task node |
|---|
|
 Image Modified Image Modified
|
Once the t-SNE task has completed, we can view the t-SNE plots
...
| Numbered figure captions |
|---|
| SubtitleText | Viewing t-SNE plot of a single sample |
|---|
| AnchorName | Viewing single-sample t-SNE |
|---|
|
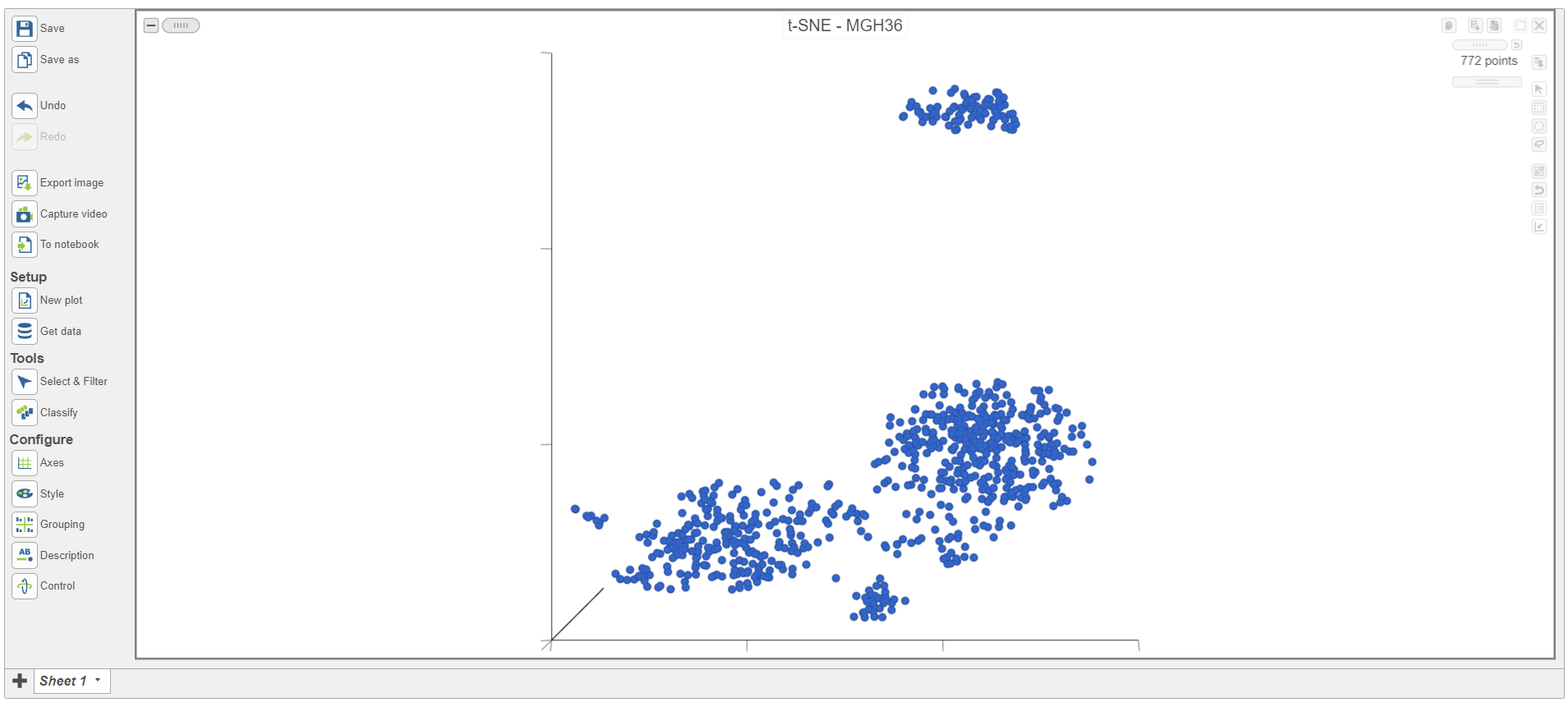 Image Modified Image Modified
|
The t-SNE plot is in 3D by default. To change the default, click your avatar in the top right > Settings > My Preferences and edit your graphics preferences and change the default scatter plot format from 3D to 2D.
...
| Numbered figure captions |
|---|
| SubtitleText | Viewing t-SNE plot of MGH42 |
|---|
| AnchorName | Viewing single-sample t-SNE (2) |
|---|
|
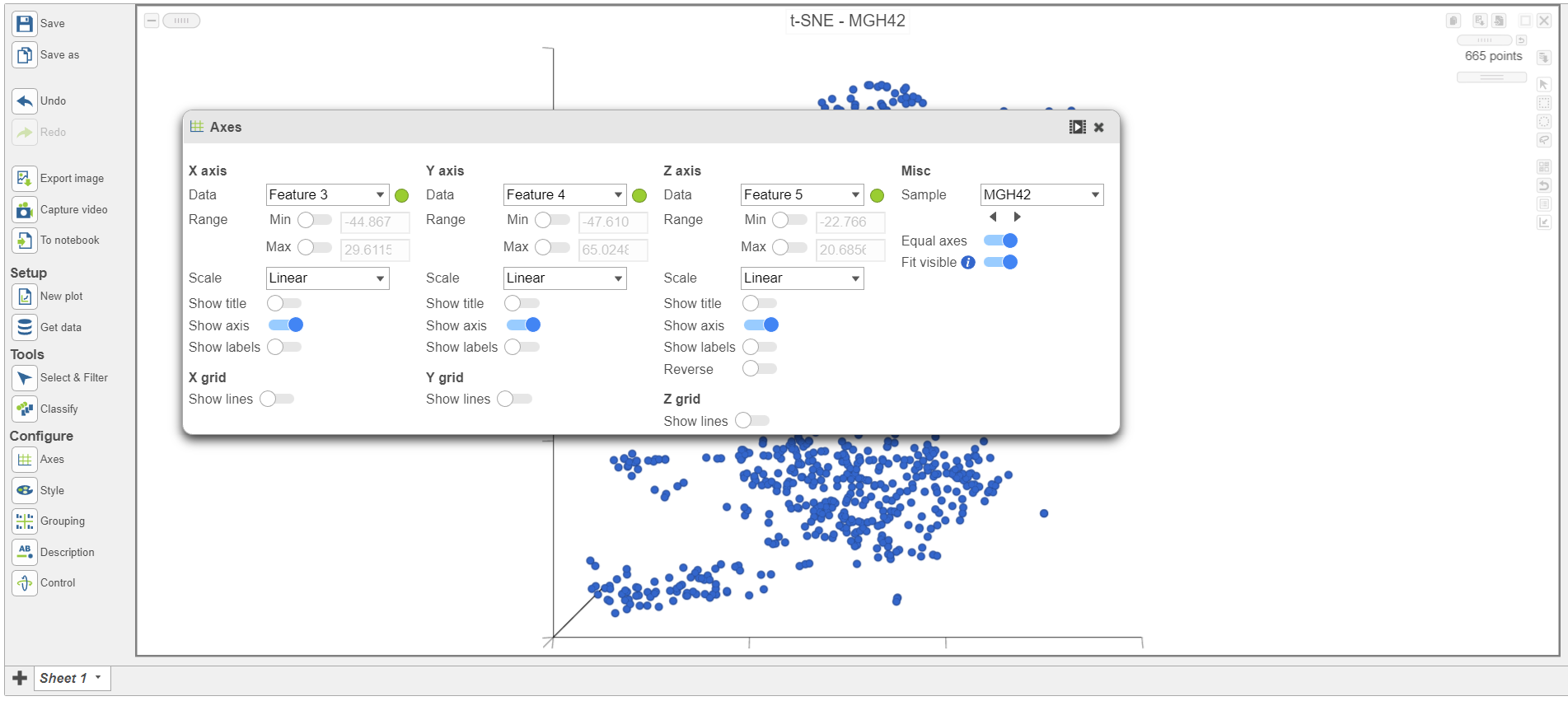 Image Modified Image Modified
|
The goal of this analysis is to compare malignant cells from two different glioma subtypes, astrocytoma and oligodendroglioma. To do this, we need to identify the malignant cells we want to include and which cells are the normal cells we want to exclude.
...
| Numbered figure captions |
|---|
| SubtitleText | Coloring cells by BCAN expression |
|---|
| AnchorName | Selecting gene |
|---|
|
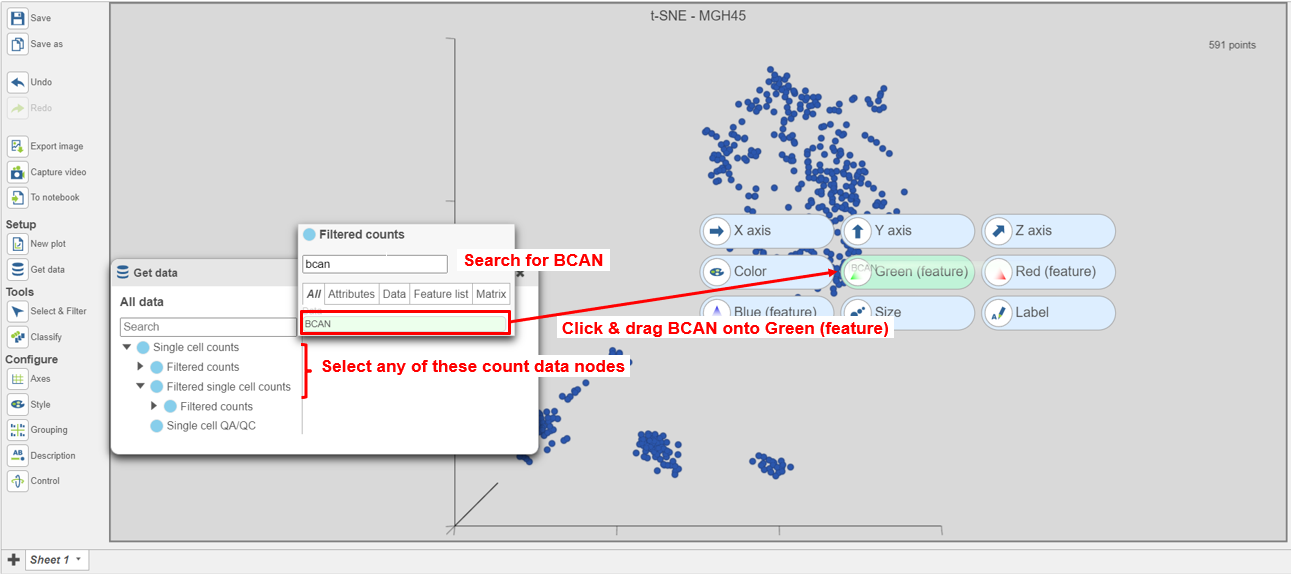 Image Modified Image Modified
|
The cells will be colored from black to green based on their expression level of BCAN, with cells expressing higher levels more green (Figure 9). BCAN is highly expressed in glioma cells.
...
| Numbered figure captions |
|---|
| SubtitleText | Classifying selection |
|---|
| AnchorName | Classifying cells |
|---|
|
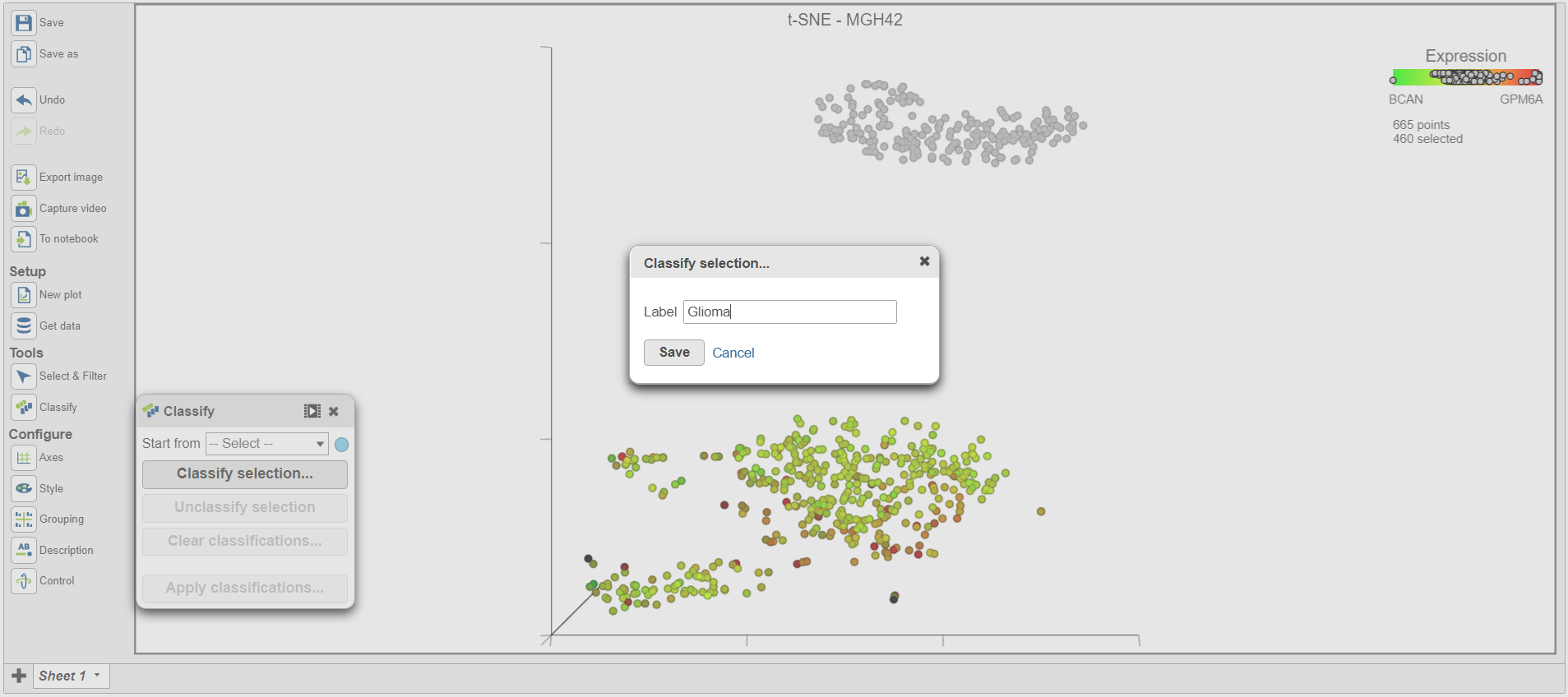 Image Modified Image Modified
|
Once cells have been classified, the classification is added to Classify. The number of cells belonging to the classification is listed. In MGH42, there are 460 glioma cells (Figure 15).
...
| Numbered figure captions |
|---|
| SubtitleText | Classifying malignant cells in sample MGH45 |
|---|
| AnchorName | Classifying cells |
|---|
|
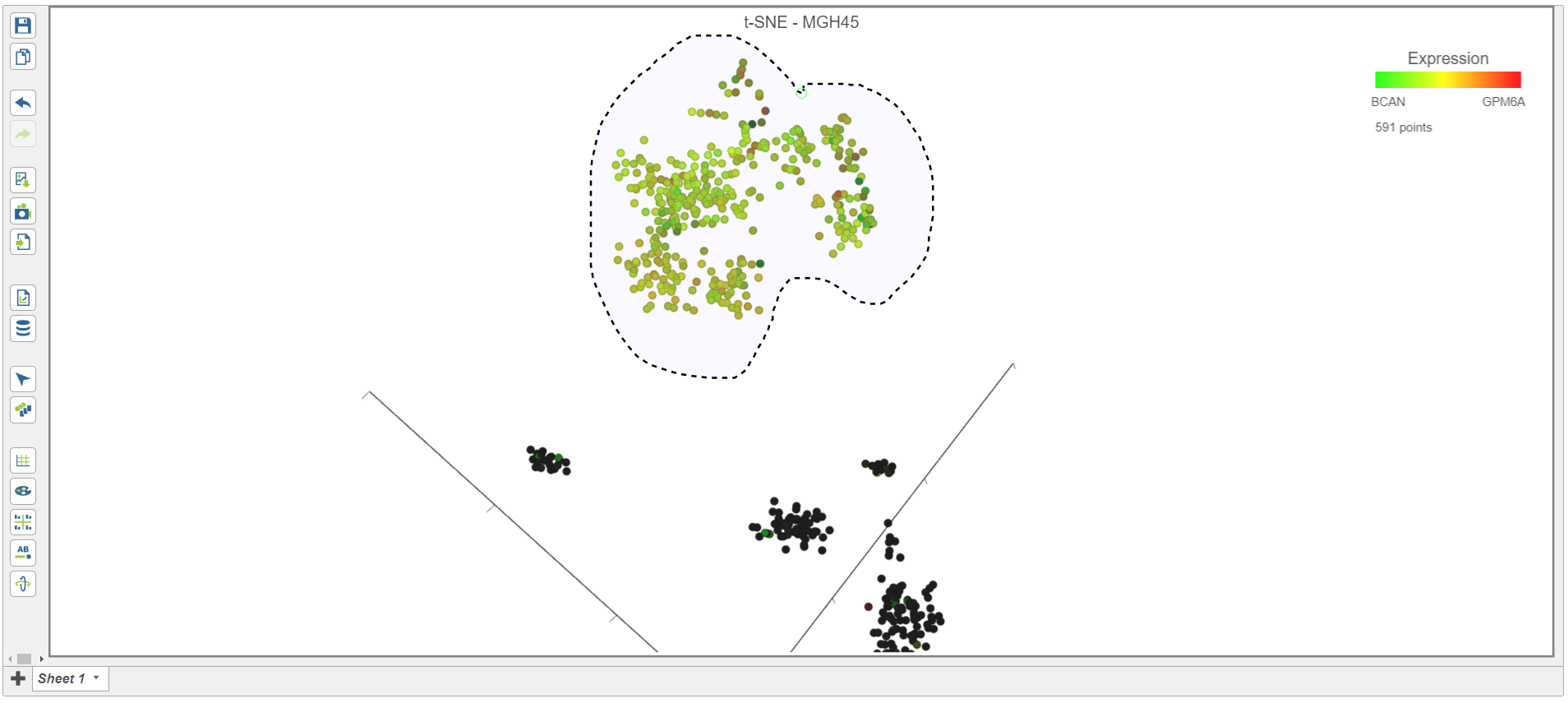 Image Modified Image Modified
|
- Select Classify selection in the Classify icon
- Type Glioma or select Glioma from the drop-down list (Figure 17)
- Click Save
...
| Numbered figure captions |
|---|
| SubtitleText | Adding cells in a second sample to an existing classification |
|---|
| AnchorName | Classifying cells as an existing classification |
|---|
|
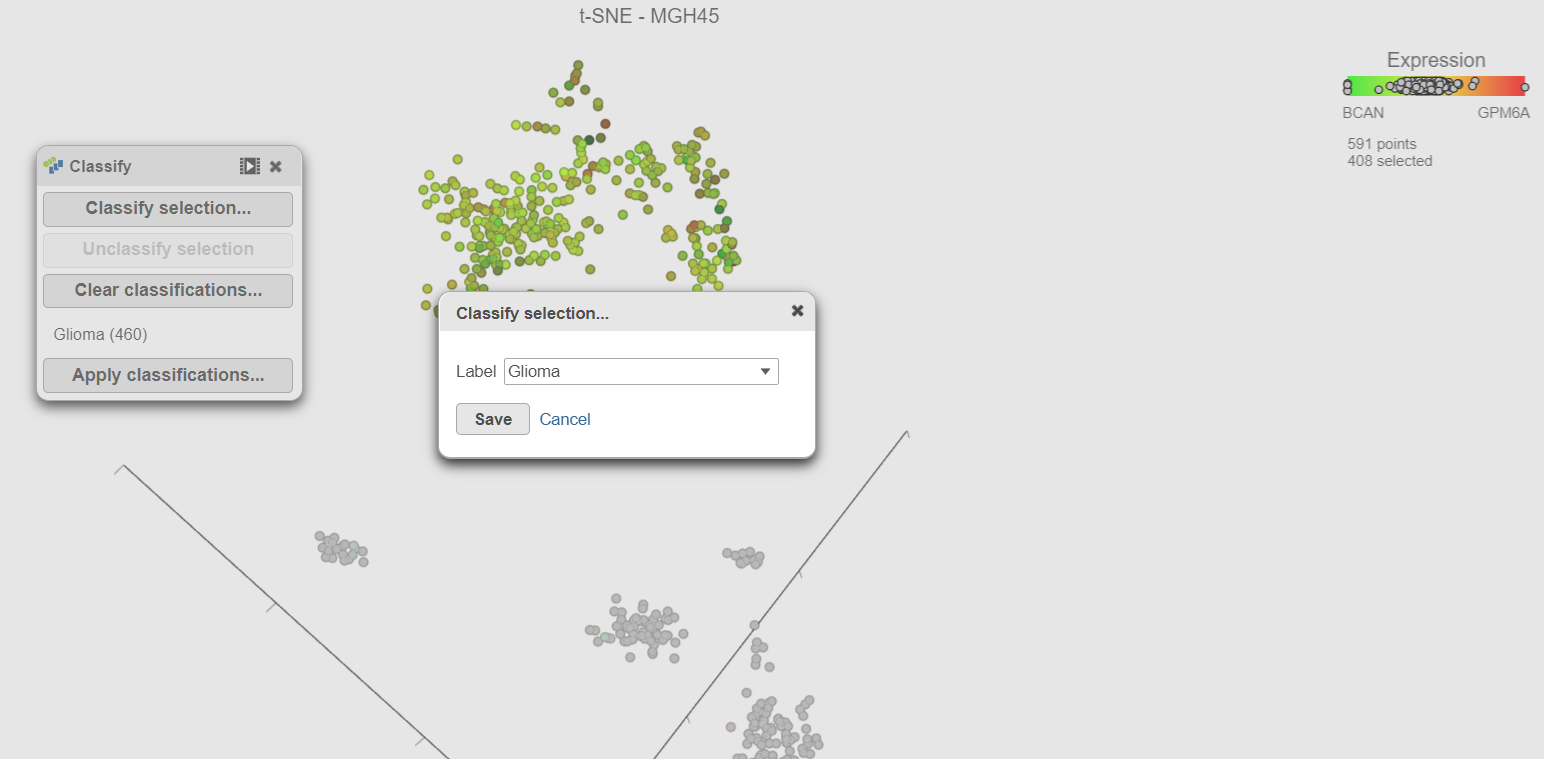 Image Modified Image Modified
|
- Repeat these steps for each of the 6 remaining samples. Remember to go back to the first sample (MGH36) to classify the glioma cells in that samples too.
...
| Numbered figure captions |
|---|
| SubtitleText | Viewing the multi-sample t-SNE plot |
|---|
| AnchorName | multi-sample t-SNE plot |
|---|
|
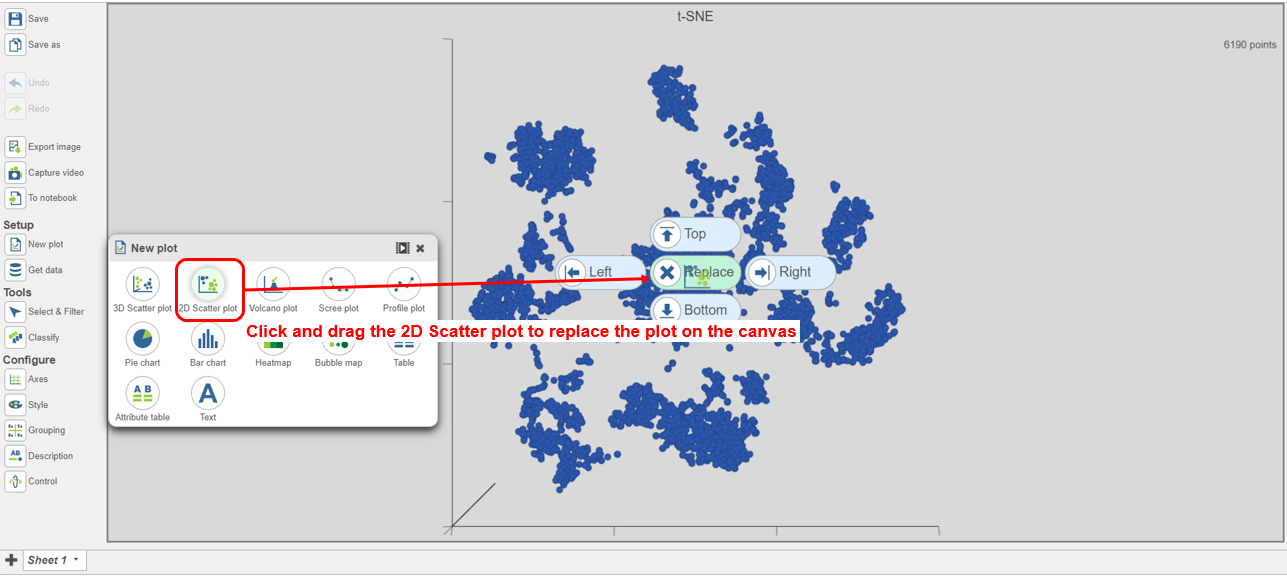 Image Modified Image Modified
|
- Search for and select green t-SNE data node (Figure 25)
...
| Numbered figure captions |
|---|
| SubtitleText | Viewing the multi-sample t-SNE plot in 2D |
|---|
| AnchorName | Viewing 2D t-SNE |
|---|
|
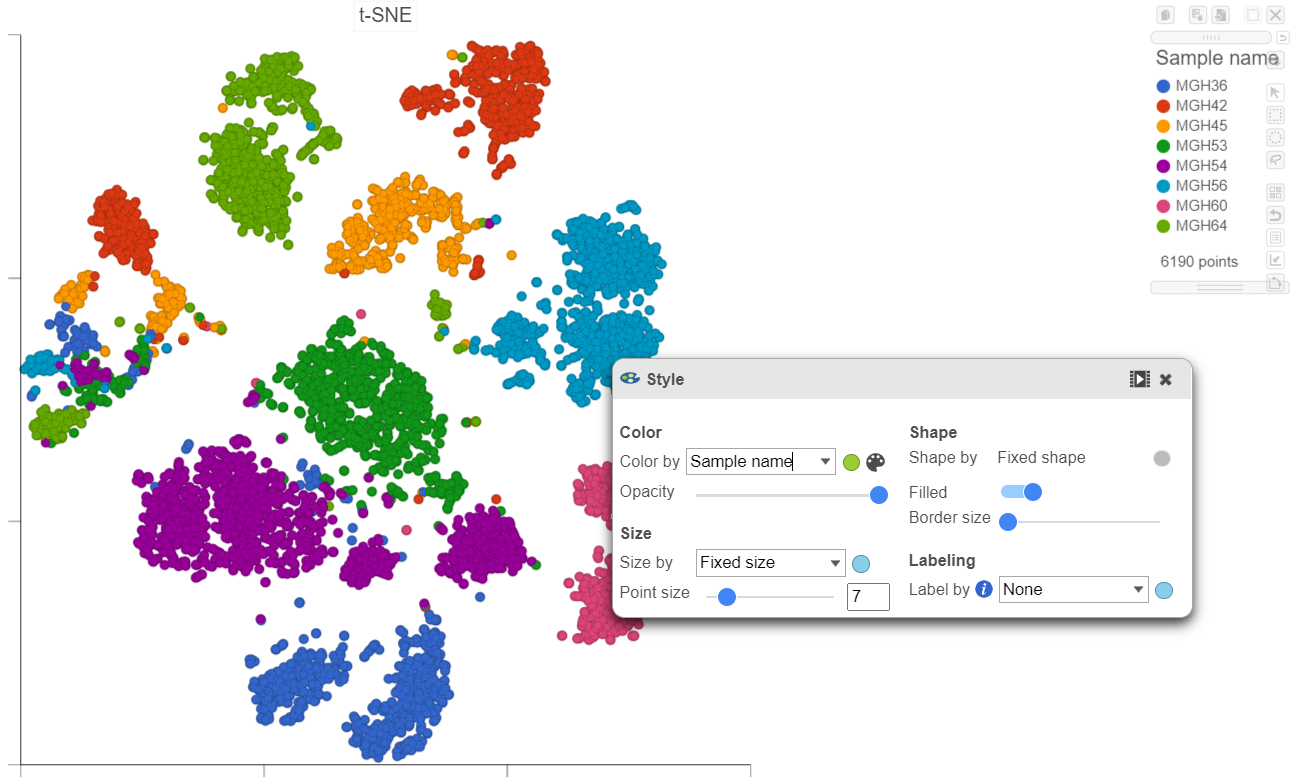 Image Modified Image Modified
|
Using marker genes, BCAN (glioma), CD14 (microglia), and MAG (oligodendrocytes), we can assess whether these multi-sample clusters belong to our known cell types.
...










