Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The t-SNE plot is in 3D by default. To change the default, click your avatar in the top right > Settings > My Preferences and edit your graphics preferences and change the default scatter plot format from 3D to 2D.
...
Each sample has its own plot. We can switch between samples.
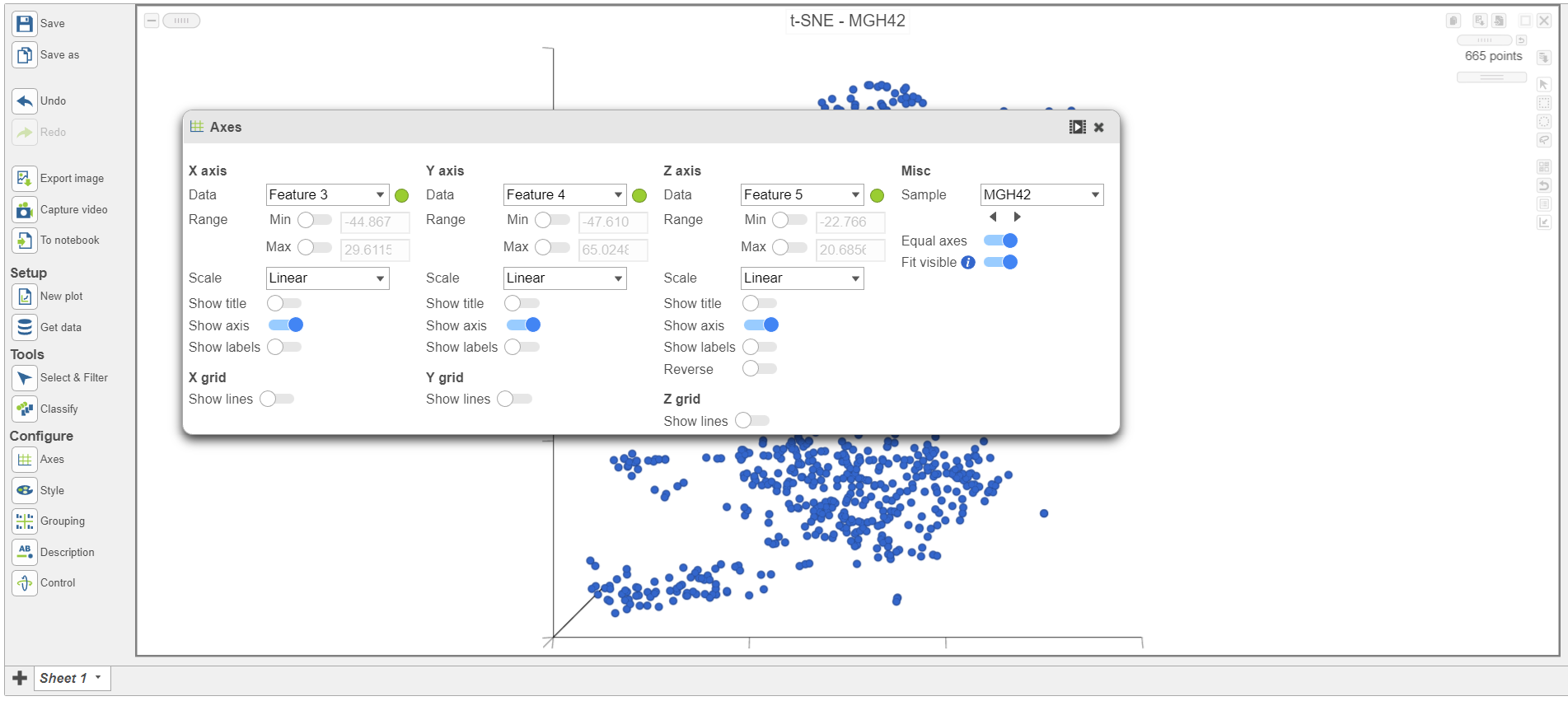
- Scroll down on the Configuration card on the left and expand the Data card Open the Axes icon on the left under Configure (Figure 7)
- Navigate to Misc
- Select the icon below the sample Sample name to go to the next sample
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The goal of this analysis is to compare malignant cells from two different glioma subtypes, astrocytoma and oligodendroglioma. To do this, we need to identify the malignant cells we want to include and which cells are the normal cells we want to exclude.
...
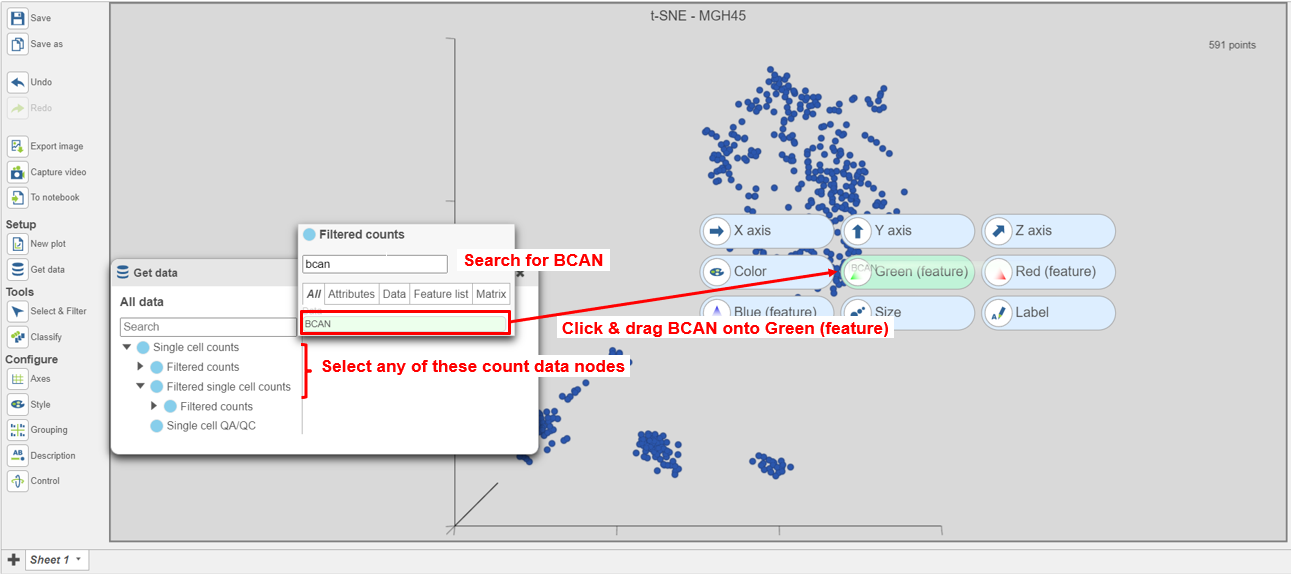
- Select any of the count data nodes from the Data card on Get data on the left (Single cell counts, or any of the Filtered counts, Figure 8)
- Search for the BCAN gene
- Click and drag the BCAN gene onto the plot and drop it over the Green (feature) option
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
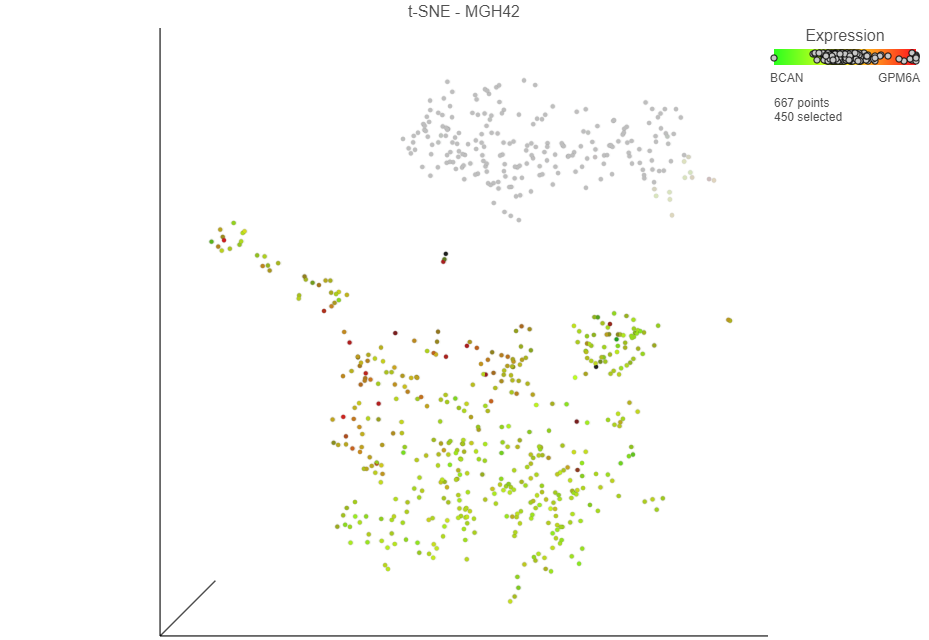
The cells will be colored from black to green based on their expression level of BCAN, with cells expressing higher levels more green (Figure 9). BCAN is highly expressed in glioma cells.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
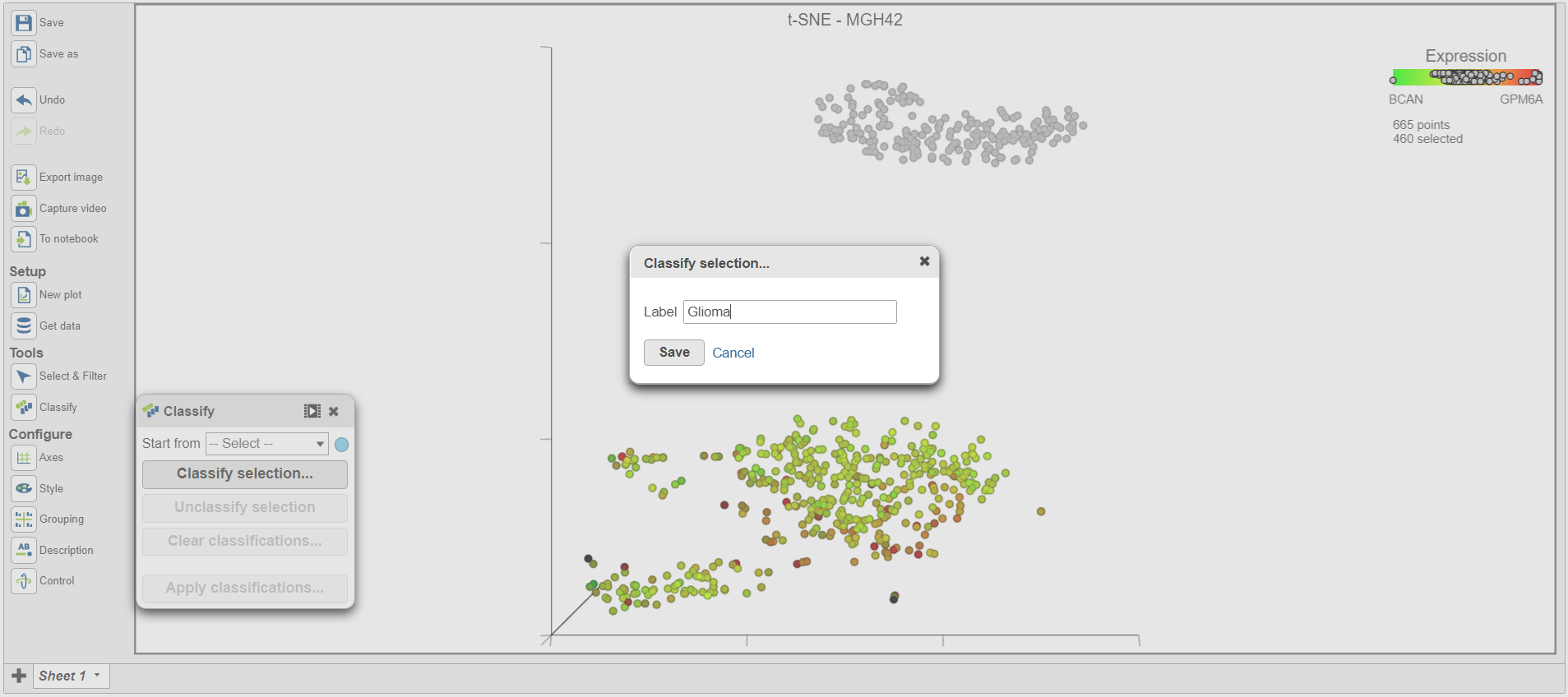
- Click Classify selection in the Classification card on the right in the Classify icon under Tools
A dialog to give the classification a name will appear.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
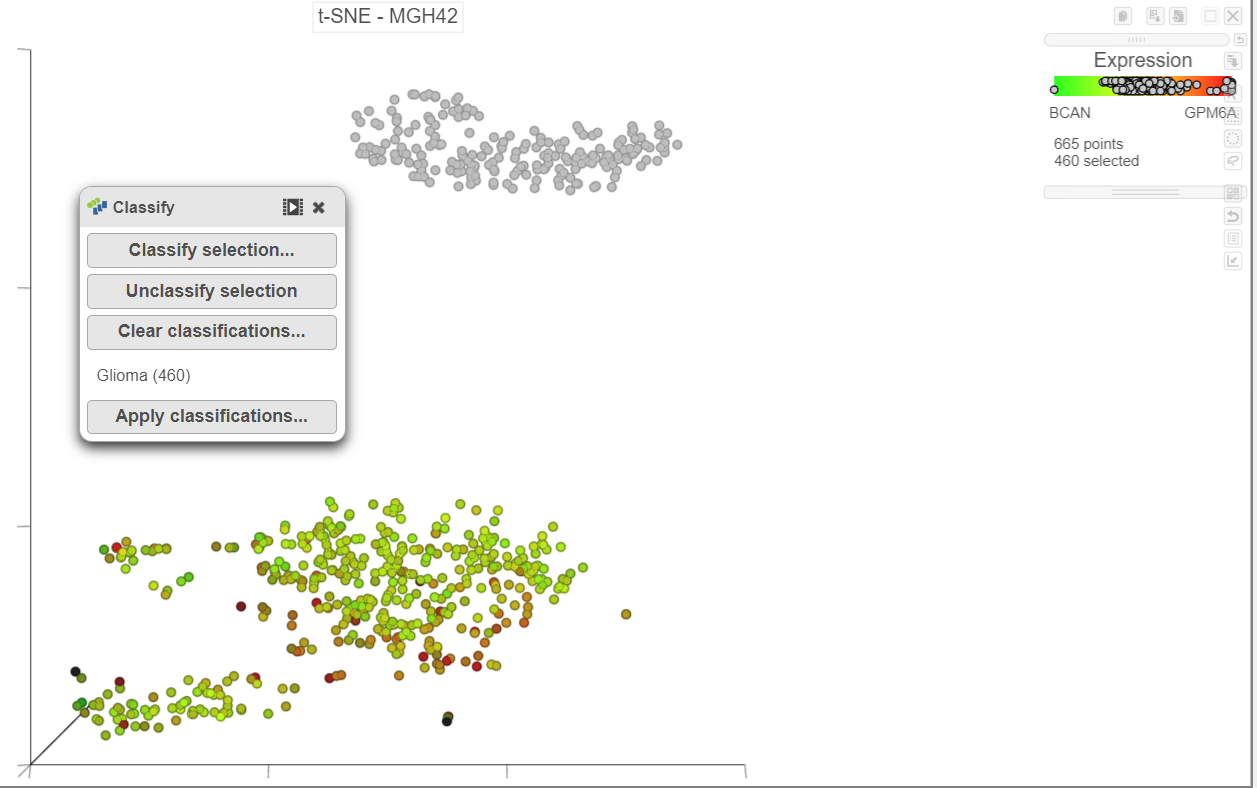
Once cells have been classified, the classification is added to the Classifications card on the right Classify. The number of cells belonging to the classification is listed. In MGH42, there are 450 460 glioma cells (Figure 15).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Classifications made on the t-SNE plot are retained as a draft as part of the data viewer session. In this tutorial, we will classify malignant cells for each sample before we save and apply the classifications, but if necessary, you can save the data viewer session by clicking the save Save icon on the left to retain all of the formatting and draft classifications. The data viewer session will be stored under the Data viewer tab and can be re-opened to continue making classifications at a later time.
- Switch to pointer mode by clicking in the top right corner of the plot
- Deselect the cells by clicking on any blank space on the plotScroll down on the Configuration card on the left and expand the Data card
- Open Axes and navigate to Sample under Misc
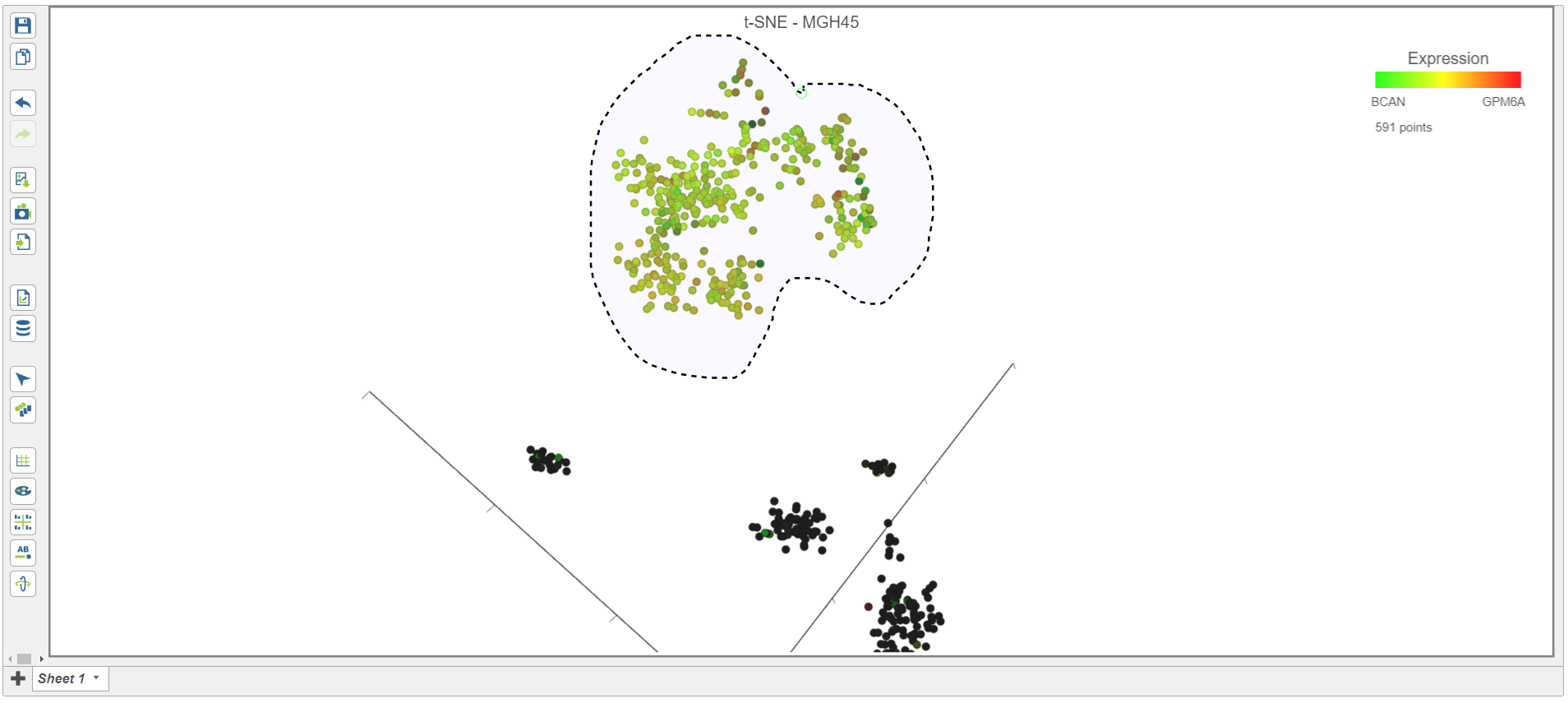
- Select the icon below the sample name to go to the next sample, MGH45
- Rotate the 3D t-SNE plot to get a better view of cells from the green, red, and yellow cluster
- Switch to lasso mode by selecting in the top right corner of the plot
- Draw the lasso around the cluster of colored cells and click the circle to close the lasso (Figure 16)
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
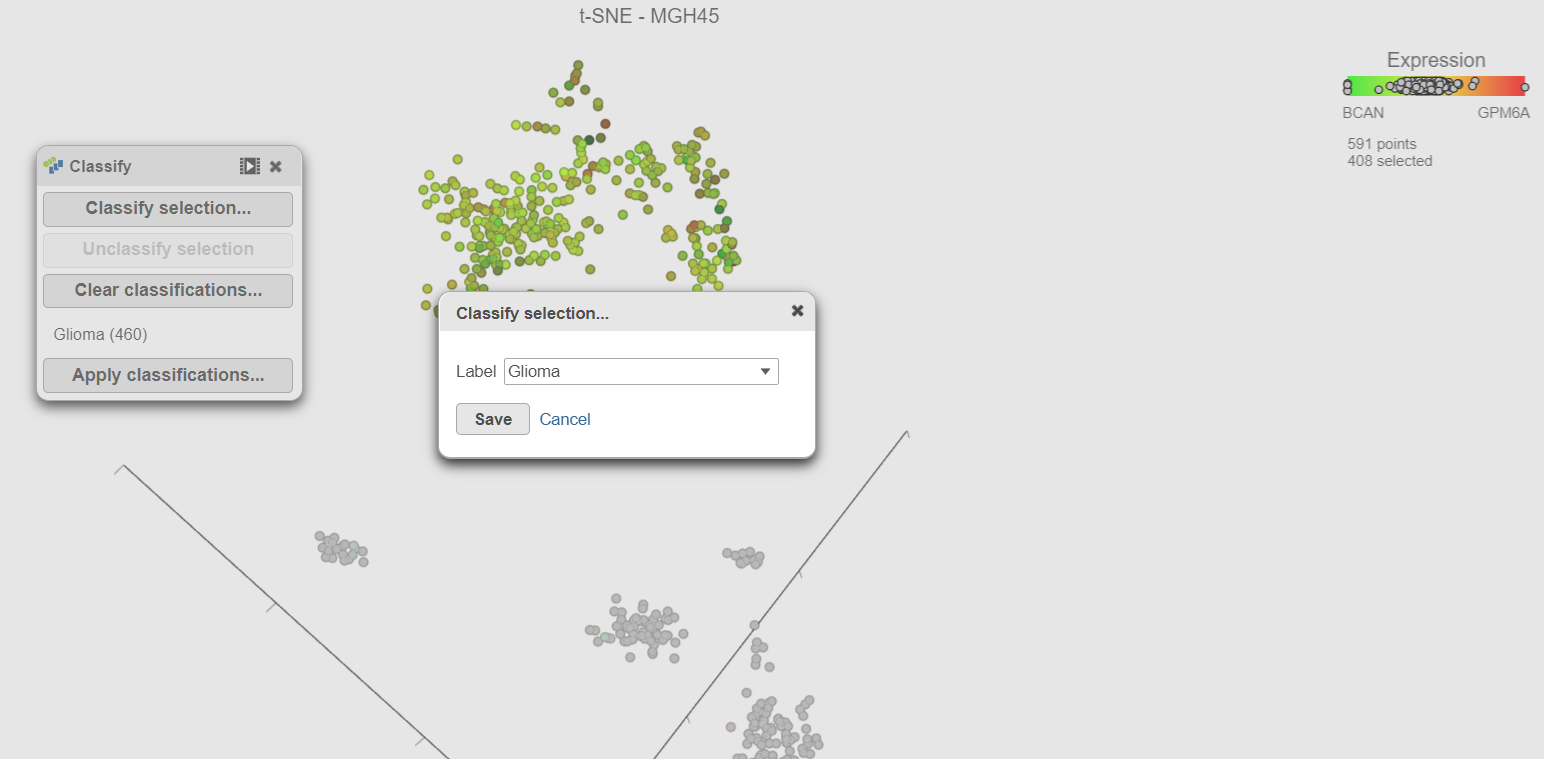
- Select Classify selection in the Selection card on the rightthe Classify icon
- Type Glioma or select Glioma from the drop-down list (Figure 17)
- Click Save
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Repeat these steps for each of the 6 remaining samples. Remember to go back to the first sample (MGH36) to classify the glioma cells in that samples too.
There should be 5,348 322 glioma cells in total across all 8 samples. With the malignant cells in every sample classified, it is time to save the classifications.
...