Page History
The Peak calling task is used to detect enriched genomic regions on reads generated from nucleic acid enrichment experiments such as ChIP-seq, DNase-seq, and MeDIP-seq. experiments. Partek® Flow® provides the widely used method of MACS2-model-based analysis1 (http://liulab.dfci.harvard.edu/MACS/) to find peaks. It can be performed with or without control sample.
MACS2 dialog
Selecting MACS2 from the context sensitive menu will bring up the MACS2 task dialog. The interface will appear differently depending on the input aligned data node and whether there are sample attributes available in the Data tab.
If the selected aligned data node was imported, the reference assembly used during data alignment needs to be specified. Choose the Assembly from the drop-down list within the MACS2 dialogue (Figure 1). If the selected aligned data node was generated by Partek Flow, this option will not appear.
While processing 10x Chromium Single Cell Multiome ATAC + Gene Expression sequencing data via ‘cellranger-arc count’ pipeline, Feature linkages analysis is performed as pairs of genomic features, such as peaks and genes, that have significant correlation in signals across cells. Because it provides a basis for inferring enhancer-gene targeting relationships and constructing transcriptional networks. The features with strong linkages are considered to be “co-expressed” and enriched for a shared regulatory mechanism.
Partek Flow provides the opportunity to our users to explore the linkage relationships among different features including peaks and genes, peaks and peaks, and genes and genes. A tab-delimited file containing information of feature linkages inferred from Flow Cell Ranger - ATAC task will be loaded into Integrative Genome Viewer (IGV)[1] for exploration if Feature linkage analysis task has been completed successfully.
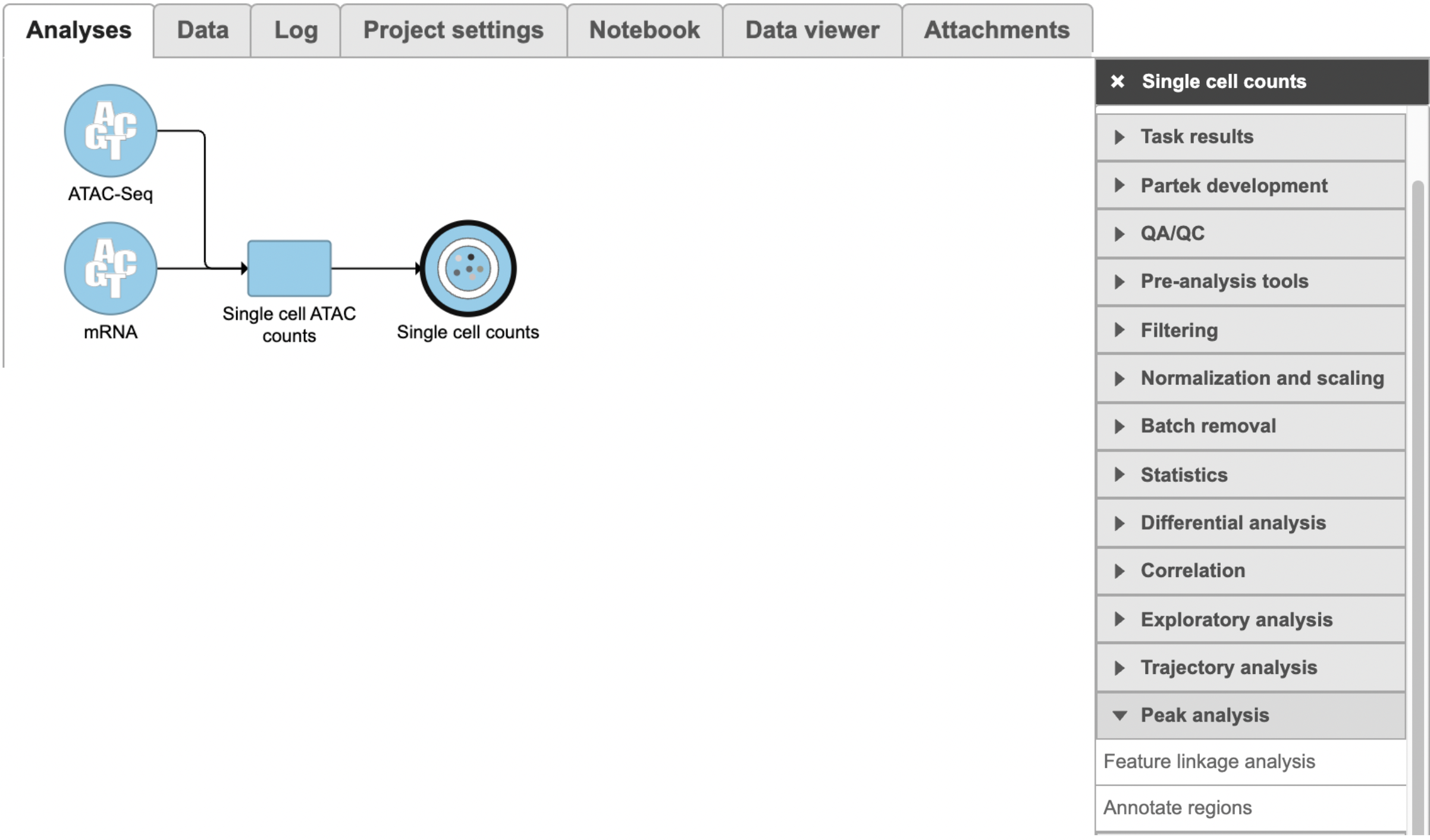
Running Feature linkage analysis
To run Feature linkage analysis task (Figure 1),
- Click one datanode that has both features of ATAC and gene expression;
- Click the Feature linkage analysis task under Peak analysis section in the toolbox;
- Click the Finish button to complete the submission.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
The Effective genome size must be configured prior to running the peak caller. It refers to the size of the genomic regions that are actually mappable. This size is smaller than the actual size of an organism's whole genome because of the presence of repetitive features. They are typically about 70%-90% of the whole genome. The MACS2 authors1 have recommended presets available for four different species. Select from the drop-down menu the preset that best describes the genome you are working with. They are as follows:
- Human (Homo sapiens) – 2.7 x 109
- Mouse (Mus musculus) – 1.87 x 109
- C. elegans – 9 x107
- Fruitfly (Drosophila melongaster) – 1.2 x 108
...
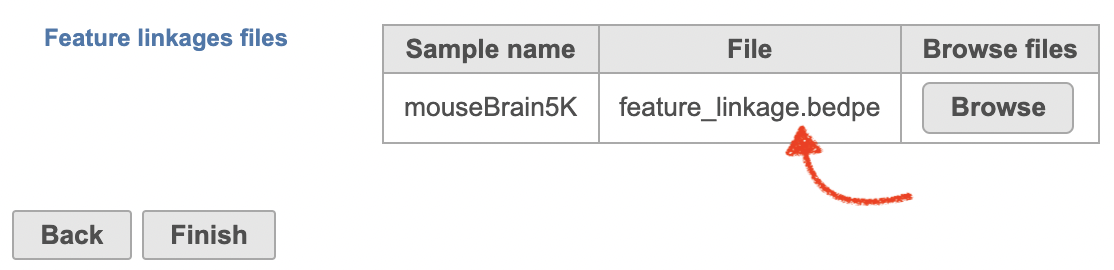
There will be no inputs needed if the FASTQ is converted to counts matrix within Flow. However, if users processed the FASTQ files outside of Partek, and imported the counts matrix into Flow later. The feature_linkage.bedpe file in outs/analysis/feature_linkage from Cell Ranger output will be needed for each sample (Figure 2) to complete the analysis.
| Numbered figure captions | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| |||||||
For data where no sample attributes are specified, the peak detection pairs need to be manually defined. In the example in Figure 1, there only two samples. Under the Define pairs section, the left panel lists all the sample names uploaded to the project (H3K27 and Mock). Add one pair at a time by dragging the corresponding samples to either the IP panel on the top-right or the Control panel on the bottom-right. If no control samples are present in the experiment, leave the Control panel blank. If more than one ChIP or Control samples are added, the samples will be combined (or pooled) during the analysis. After defining a pair, click the Add pair button.
...