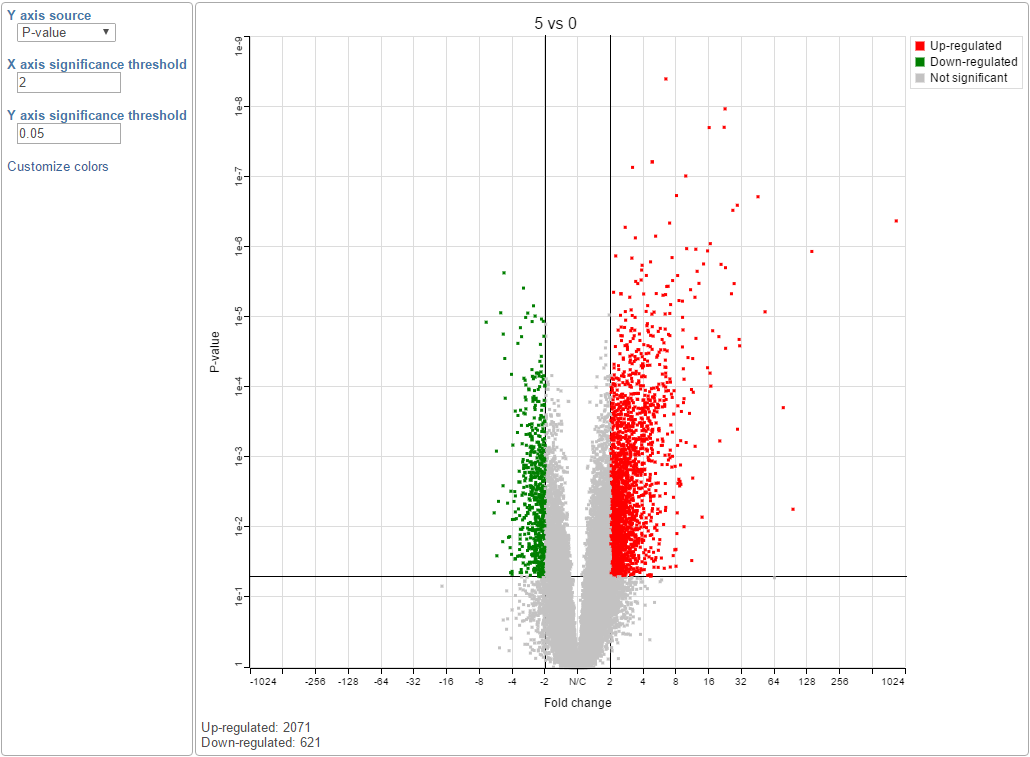
The volcano plot is a special 2-D scatter plot used to visualize significance and the magnitude of changes in features (e.g. genes or transcripts) within a given comparison. By convention, the X-axis represents the fold change between the two groups and is on a log2 scale. On the other hand, the Y-axis shows negative log10 of the p-values from the statistical test of the comparison and it is on a log10 scale.
As a prerequisite for invoking the volcano plot, you must run a Differential gene expression analysis. When setting-up the analysis, include all relevant contrasts you would like to view the volcano plots for.
...
| Numbered figure captions |
|---|
| SubtitleText | Invoking a volcano plot from a Feature list data node |
|---|
| AnchorName | invoke-volcano-plot |
|---|
|
|
Each point on the plot represents the statistical result for a single
feature (e.g gene, transcript
or featureetc). The
color of the point reflects fold change: genes with a fold change above 2 (up-regulated) are in green, genes with a fold change below -2 (down-regulated) are in red. The remaining genes are in grey (Figure 2). The plot header is derived from the name of the contrast.Thick vertical lines representing black vertical and horizontal lines represent threshold of fold change and p-value respectively. By default the two vertical lines represent fold changes of -2 and +2
are highlighted. A thick , the horizontal line represents
a significant p-value of 0.05. The number of
genes features that are up/
downregulated down regulated by at least 2
fold and have a p-value less than 0.05 are
listed at the
upper-right and lower
-left corner of the plot
, they are highlighted in different colors.
By default significantly up regulated features are in red, significantly down regulated features are in green. The remaining features are in grey (Figure 2). The plot header is derived from the name of the contrast.You can manually configure the Y axis source from the drop-down list and change the threshold of p-value and fold change, then the position of the lines and number of features in the significant areas will be updated accordingly. Click on the Customize colors can manually change the color of the dots (features) in the different areas.
| Numbered figure captions |
|---|
| SubtitleText | Volcano plot: each dot on the plot is a single gene/transcript/feature. Horizontal axis: fold change (in log2 scale); vertical axis: p-value (in log10 scale). Colour coding is based on the fold change. Thick vertical lines highlight fold changes of -2 and +2, while a thick horizontal line represents a p-value of 0.05 |
|---|
| AnchorName | volcano-plot |
|---|
|
|
...
