Page History
...
If Partek® Flow® has no information on the genome build, you will need to provide the species and genome build in a subsequent dialog (not shown). Otherwise, chromosome view will come up directly.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
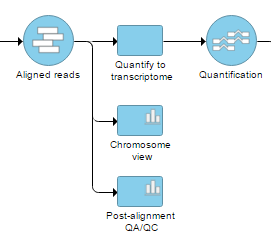
A new Chromosome view task node will be added to the canvas (Figure 2) and in order to invoke the viewer <double-click> on the node (you can also select it and then go to Task report in the menu). When invoked in this way, the default visualization in the Chromosome view is the first 100,000 bases of the first chromosome.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
|
| Page Turner | ||
|---|---|---|
|
| Rate Macro | ||
|---|---|---|
|
...
Overview
Content Tools