Page History
...
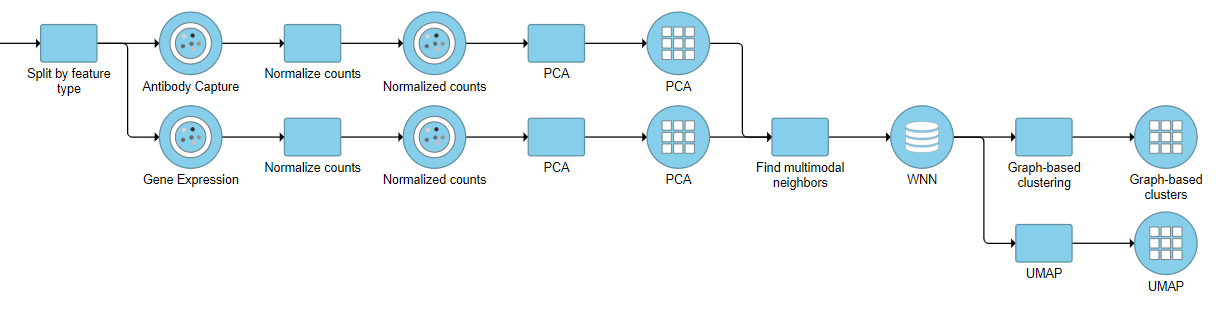
Multi-omics single cell analysis is based on simultaneous detection of different types of biological molecules on the same cells. Common multi-omics techniques include feature barcoding or CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) technologies, which enable parallel assessment of gene and protein expression. Specific bioinformatics tools have been developed to enable scientists to integrate results of multiple assays and learn relative importance of each type (or each biological molecule) in identification of cell types. Partek Flow supports weighted nearest neighbour neighbor (WNN) analysis (1), which can help combine output of two molecular assays.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
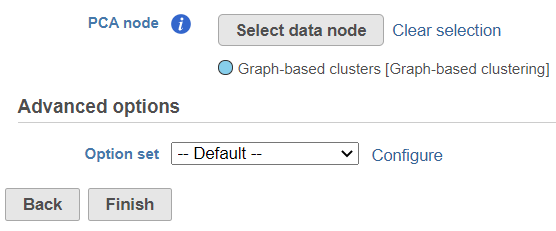
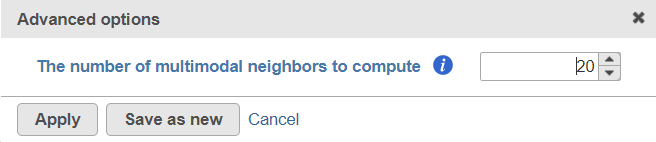
To customize the Advanced options, select the Configure link (Figure 3). At present you can only change the number of nearest neighbors for each modality (-k.nn option of the Seurat package); the default value is 20 (Figure 4). An illustration on how to use that option to assess the robustness of WNN analysis , please see can be found in Hao et al. (1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
For an excellent illustration on advantages of WNN algorithm for identification of cell types, please see this blog post.
...