...
| Numbered figure captions |
|---|
| SubtitleText | Quantify to annotation model(Partek E/M) dialog |
|---|
| AnchorName | em |
|---|
|
 Image Removed Image Removed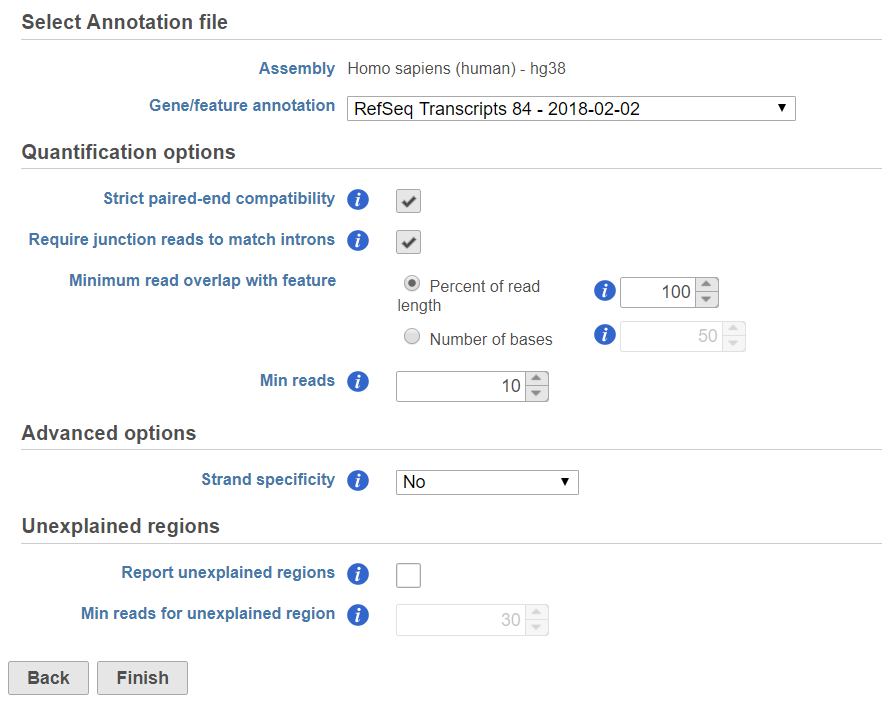 Image Added Image Added
|
If the bam file is imported, you need to select the assembly with which the reads were aligned to, and which annotation model file you will use to quantify from the drop-down menus (Figure 2).
...
| Numbered figure captions |
|---|
| SubtitleText | Specify the genome assembly with which the bam files are generated from and transcriptome annotation from the drop-down menu |
|---|
| AnchorName | specify-annot |
|---|
|
 Image Removed Image Removed Image Added Image Added
|
In the Quantification options section, when the Strict paired-end compatibility check button is selected, paired end reads will be considered compatible with a transcript only if both ends are compatible with the transcript. If it is not selected, reads with only one end have alignment that is compatible with the transcript will also be counted for the transcript ....
| Numbered figure captions |
|---|
| SubtitleText | Illustration of the three types of strand specific assays on paired end reads. _R1 and _R2 means read first-in pair and second-in-pair respectively. Arrows indicate strand directions. |
|---|
| AnchorName | strand-types |
|---|
|
 Image Removed Image Removed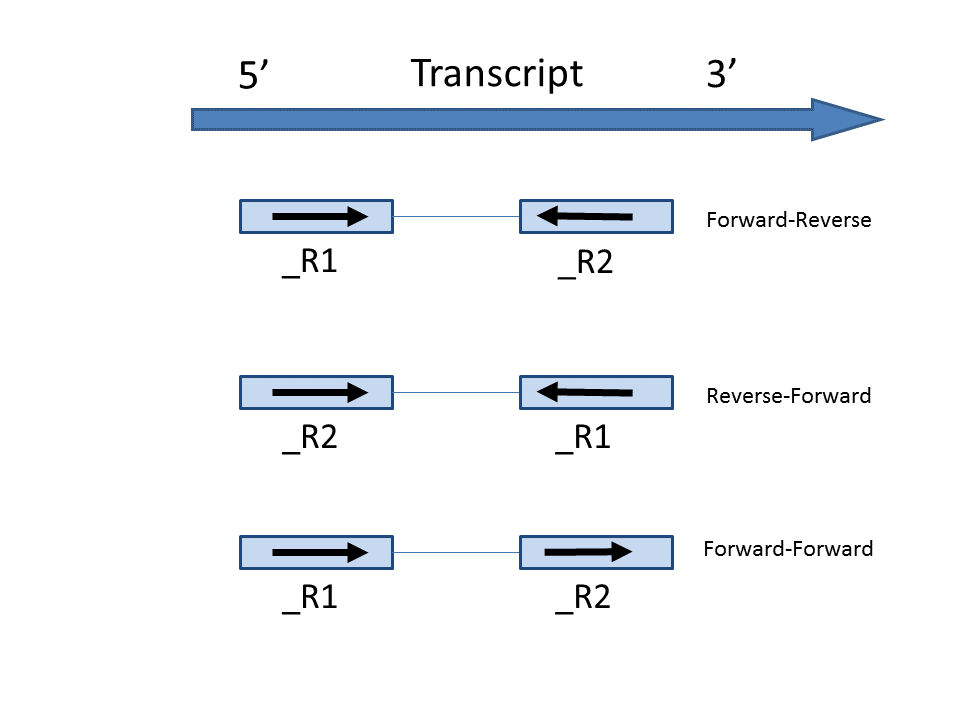 Image Added Image Added
|
Minimum read overlap with feature can be specified in percentage of read length or number of bases. By default, a read has to be 100% within a feature. You can allow some overhanging bases outside the exonic region by modifying these parameters....
| Numbered figure captions |
|---|
| SubtitleText | Summary of raw reads mapping to genes based on the RefSeq annotation file provided. Note that the Gene-level tab is selected. |
|---|
| AnchorName | reads-summary-em |
|---|
|
 Image Removed Image Removed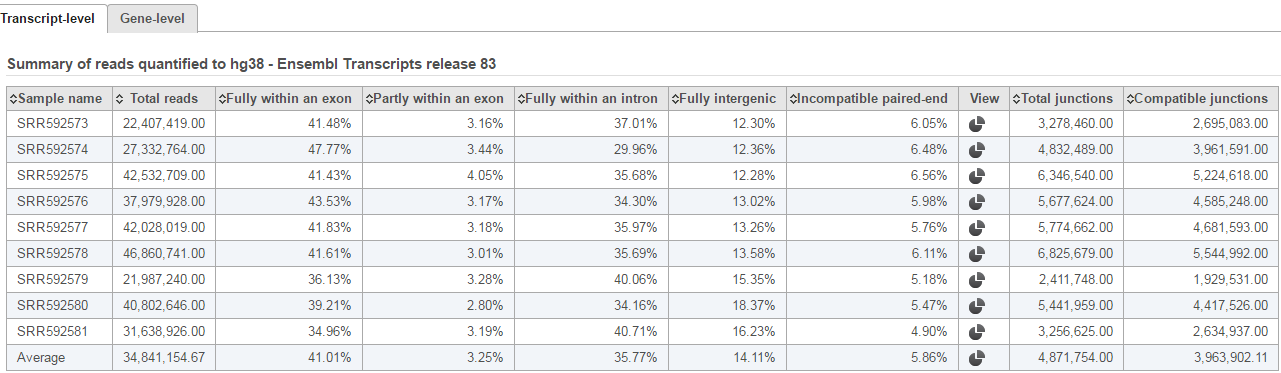 Image Added Image Added
|
The second table contains feature distribution information on each sample and across all the samples, number of features in the annotation model is displayed on the table title (Figure 5).
...
| Numbered figure captions |
|---|
| SubtitleText | Summary of feature distribution statistics |
|---|
| AnchorName | feature-dist-em |
|---|
|
 Image Removed Image Removed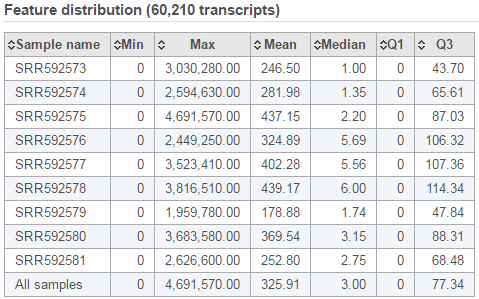 Image Added Image Added
|
The bar chart displaying the distribution of raw read counts is helpful in assessing the expression level distribution within each sample. The X-axis is the read count range, Y axis is the number of features within the range, each bar is a sample. Hovering your mouse over the bar displays the following information (Figure 6):
...
| Numbered figure captions |
|---|
| SubtitleText | Bar chart on distribution of raw read counts in each sample |
|---|
| AnchorName | bar-chart |
|---|
|
 Image Removed Image Removed Image Added Image Added
|
The coverage breakdown bar chart is a graphical representation of the reads summary table for each sample (Figure 7)
...
| Numbered figure captions |
|---|
| SubtitleText | Coverage breakdown bar chart, it is a graphical presentation of summary table on raw reads mapping to transcription based on the annotation file provided |
|---|
| AnchorName | cov-breakdown |
|---|
|
 Image Removed Image Removed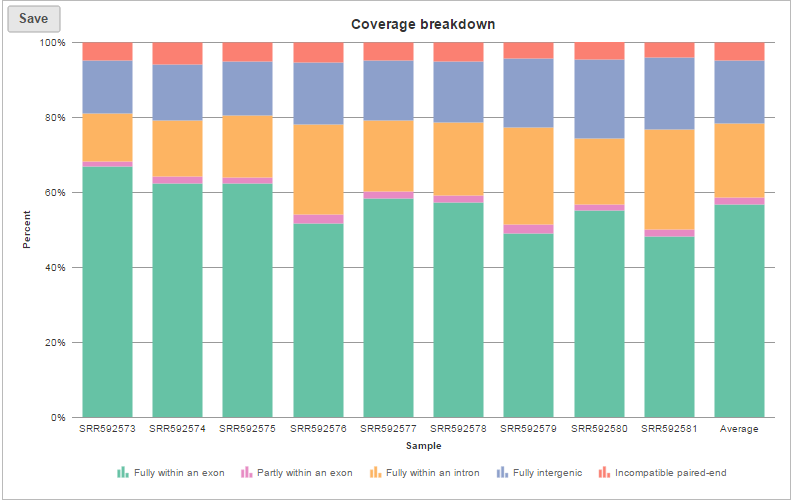 Image Added Image Added
|
In the box-whisker plot, each box is a sample on X-axis, the box represents 25th and 75th percentile, the whiskers represent 10th and 90th percentile, Y-axis represents the read counts, when you hover over each box, detailed sample information is displayed (Figure 8).
| Numbered figure captions |
|---|
| SubtitleText | Box-whisker plot on read count distribution in each sample, when mouse over a box, detailed information on the box is displayed. |
|---|
| AnchorName | box-whisker-plot |
|---|
|
 Image Removed Image Removed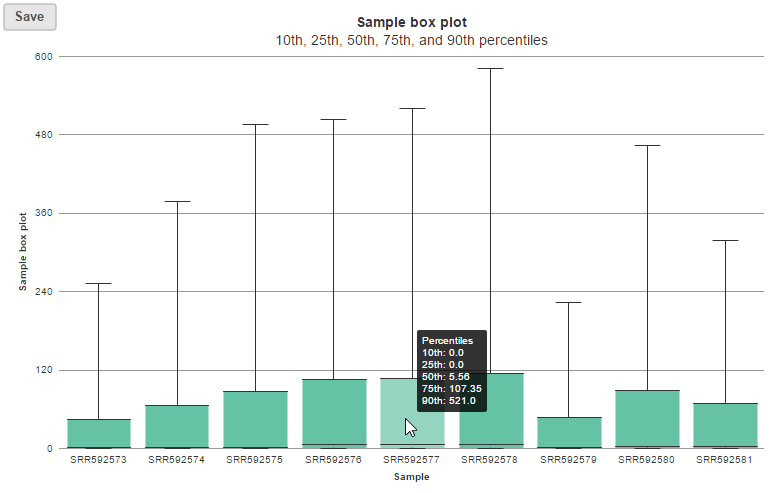 Image Added Image Added
|
In sample histogram, each line represents a sample and the range of read counts are divided into 20 bins. Clicking on a sample in the legend will hide the line for that specific sample. Hovering over each circle displays detailed information about the sample and that specific bin (Figure 9). The information includes:...
| Numbered figure captions |
|---|
| SubtitleText | Sample histogram plot, when mouse over each circle, detailed information is displayed |
|---|
| AnchorName | histogram |
|---|
|
 Image Removed Image Removed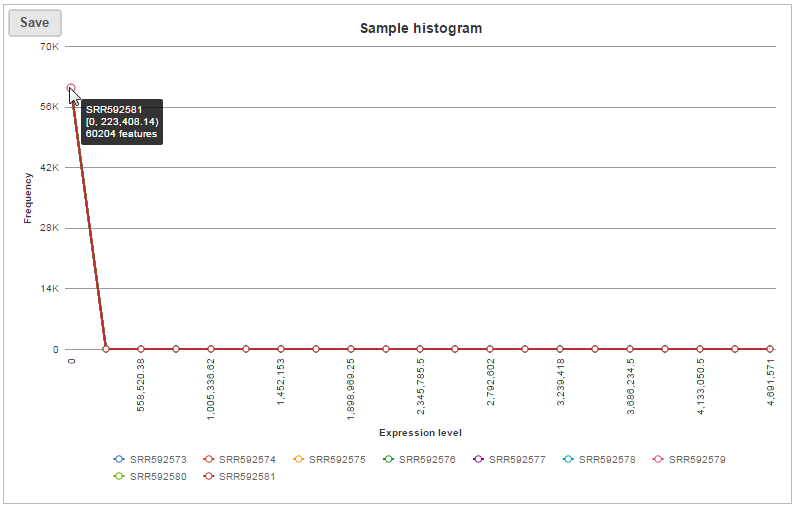 Image Added Image Added
|
The box whisker and sample histogram plots are helpful for understanding the expression level distribution across samples. This may indicate that normalization between samples might be needed prior to downstream analysis....
| Numbered figure captions |
|---|
| SubtitleText | Quantify to reference dialog |
|---|
| AnchorName | quant-reference |
|---|
|
 Image Modified Image Modified
|
- Min coverage: will filter out any features (sequence names) that have fewer reads across all samples than the value specified
...








