Page History
...
Kraken is a taxanomic sequence classifier that assigns taxonomic labels to short DNA sequences, typically from microbiome or metagenomic studies [(1]). Kraken classifies reads to a best-match location in a taxonomic tree (the lowest common ancestor), so not all sequences will be classified to a particular level such as species. Kraken matches k-mers (nucleotide sequences of k bases in length) within a read to a database of k-mer sequences from known genomes with established taxonomic relationships to perform its classifications. Partek Flow currently wraps Kraken version 2 (1).
Running Kraken
Kraken takes FASTQ files as input. Reads can be single- or paired-end.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Kraken generates a Classified sequence data nodea Taxonomic data node. This data type is the can be used as input for the the Alpha & beta diversity task.Beta diversity and Choose taxonomic level tasks. If you want to obtain the Kraken output files, select the new data node and choose Download data from the toolbox on the right.
Kraken parameters
Database name
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
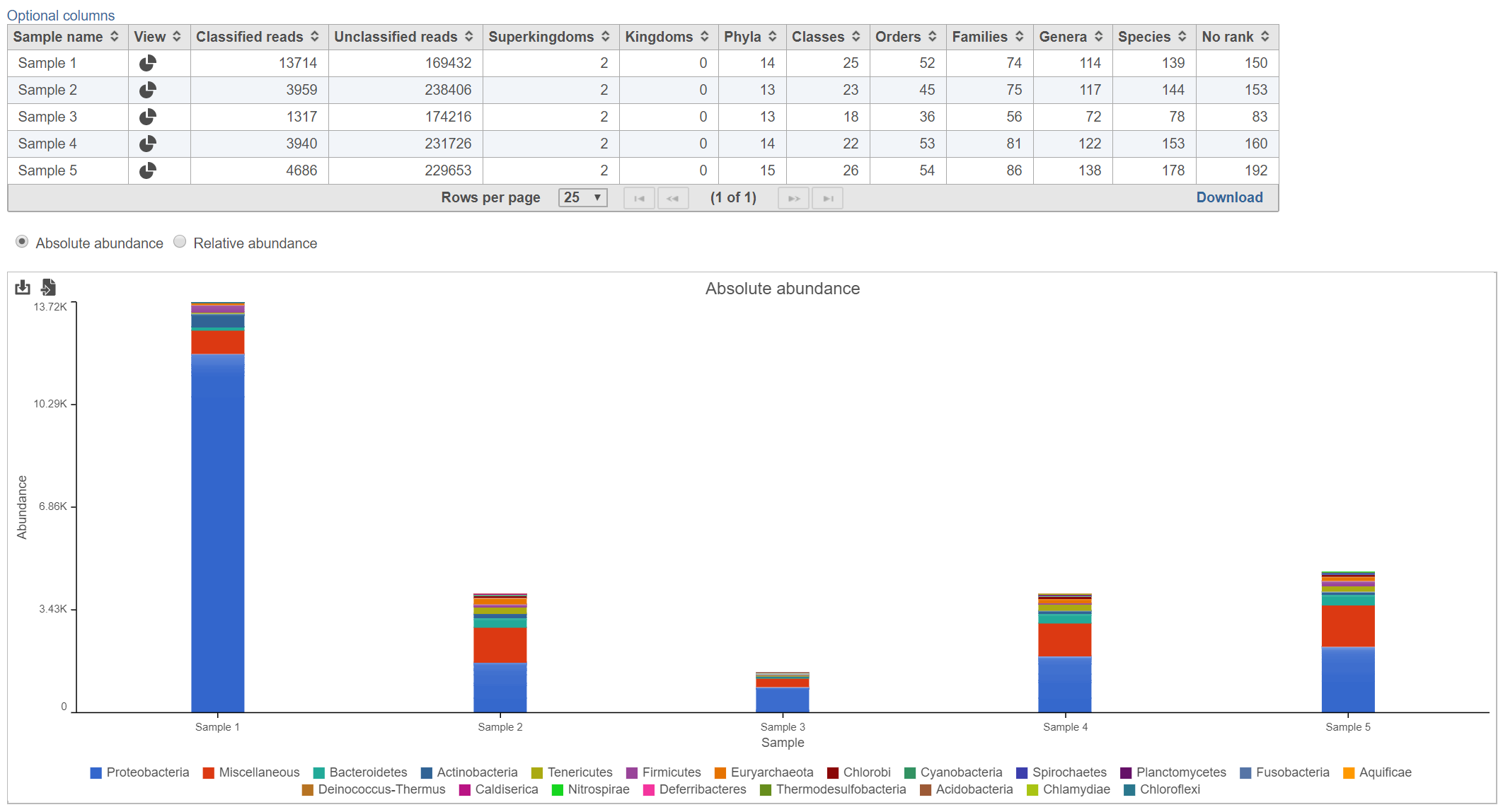
Table
The table lists each sample and gives the following values:
...
The mini-map on the upper left is shaded in green to indicate which section of the original pie chart is currently shown (Figure 5).
Downloading count data
To download a text file containing the number of classified reads for each taxon at a given taxonomic level, for each sample:
- Under the Analyses tab, click the Classified sequence data node
- Click Download data from the menu on the right
- Click the Count matrix radio button
- Select a taxonomic level
- Click Download (Figure 6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A tab-delimited text file will be downloaded to your local machine through the web browser. Taxa are on rows and samples are on columns. The values are the raw number of classified reads for each taxon, for each sample.
References
[1] Wood . Wood DE, Salzberg SL: Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology 2014, 15:R46.
2. Wood DE, Lu J & Langmead B: Improved metagenomic analysis with Kraken 2. Genome Biol 2019, 20:257
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...