...
| Numbered figure captions |
|---|
| SubtitleText | Use the Configuration panel to configure the dot plot |
|---|
| AnchorName | Configure dot plot |
|---|
|

|
- Click the project name to return to the Analyses tab
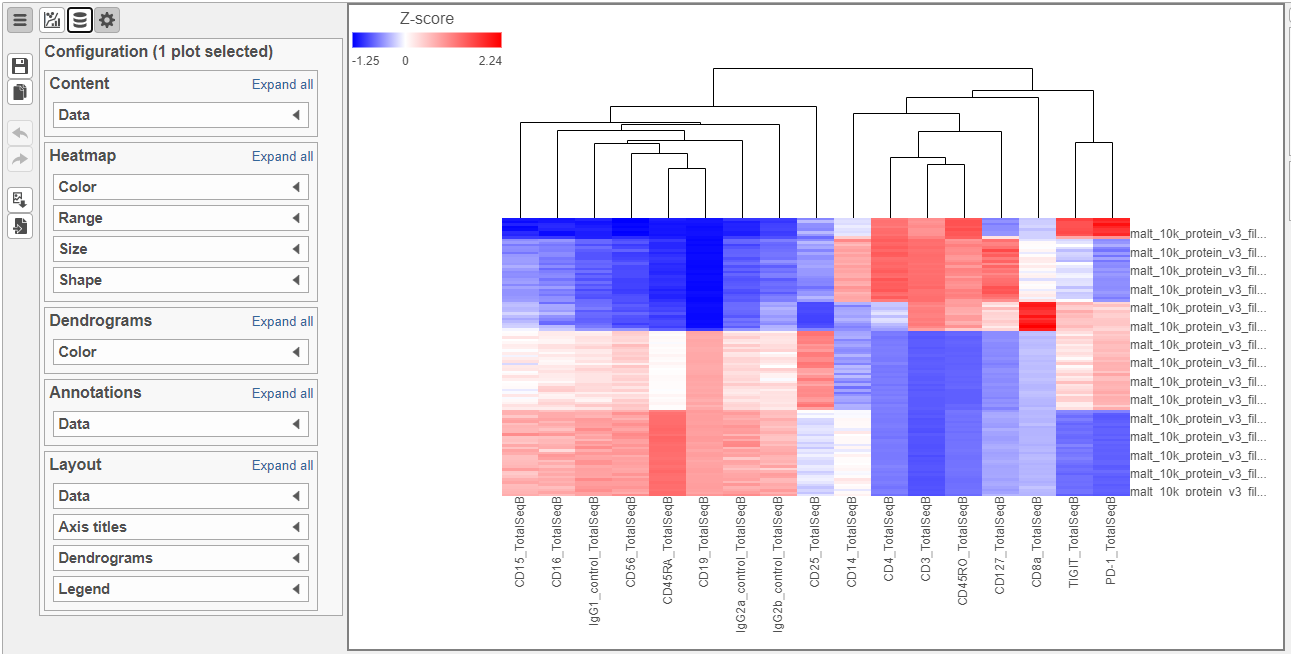
To visualize all of the proteins at the same time, we can make a hierarchical clustering heat map.
- Click the GSA data node
- Click Exploratory analysis in the toolbox
- Click Hierarchical clustering/heat map
- Check Samples at the top to cluster the cells in addition to features
- Click Finish to run with other default settings
- Double-click the Hierarchical clustering task node to open the heat map (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Heatmap showing expression of protein markers before configuration |
|---|
| AnchorName | Heatmap of proteins |
|---|
|
|
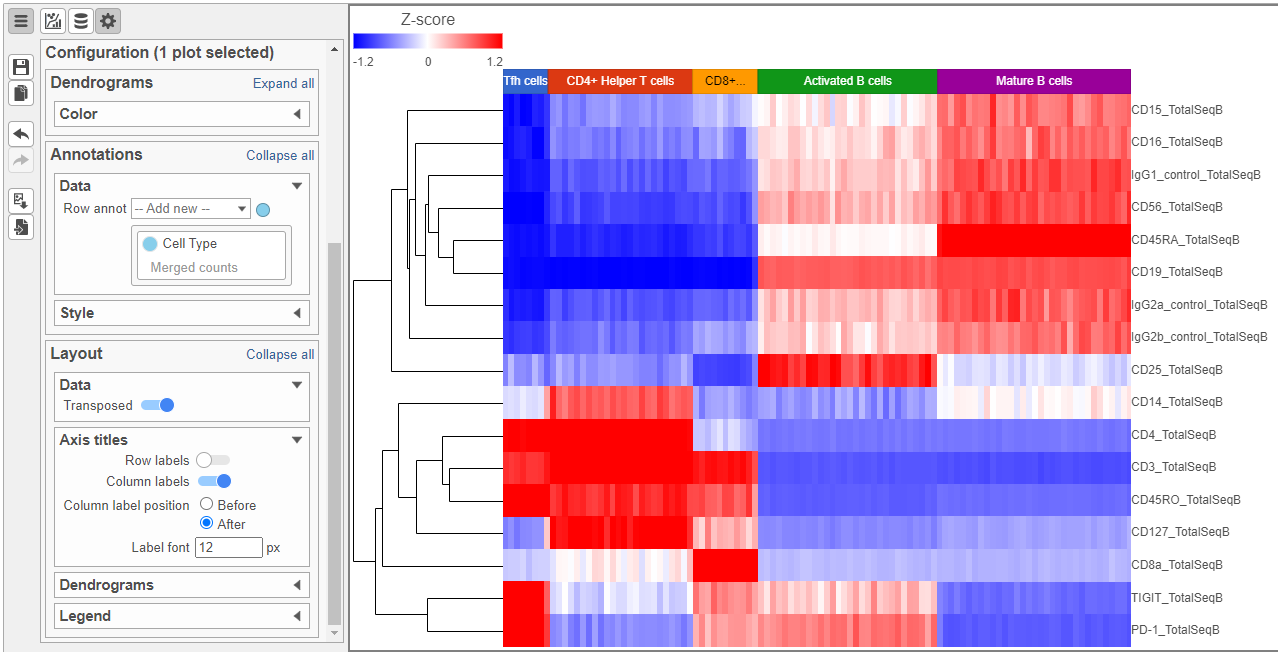
The heat map can easily be customized to illustrate our results.
- Click
 Image Modified to transpose the heat map
Image Modified to transpose the heat map - Set High to 2.6 to match the low range
- Set the Sample dendrogram to By sample attribute Cell type
- Set Attributes to Cell type
- Click
 Image Modified and set Rotation to 0
Image Modified and set Rotation to 0 - Uncheck Samples under Show labels
| Numbered figure captions |
|---|
| SubtitleText | Heatmap showing expression of protein markers after configuration |
|---|
| AnchorName | Heatmap of proteins configured |
|---|
|
|
...
Differential Analysis, Visualization, and Pathway analysis - Gene Expression Data
We can use a similar approach to analyze the gene expression data.
- Click the project name to return to the Analyses tab
- Click the Gene Expression data node
- Click Differential analysis
- Click GSA
- Check Cell type to include it in the statistical test
- Click Next
- Check Activated B cells in the top panel
- Check Mature B cells in the bottom panel
- Click Add comparison
- Click Finish to run the statistical test
As before, this will generate a GSA task node and a GSA data node.
- Double-click the GSA task node to open the task report (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | GSA report for the gene expression data |
|---|
| AnchorName | GSA genes result |
|---|
|
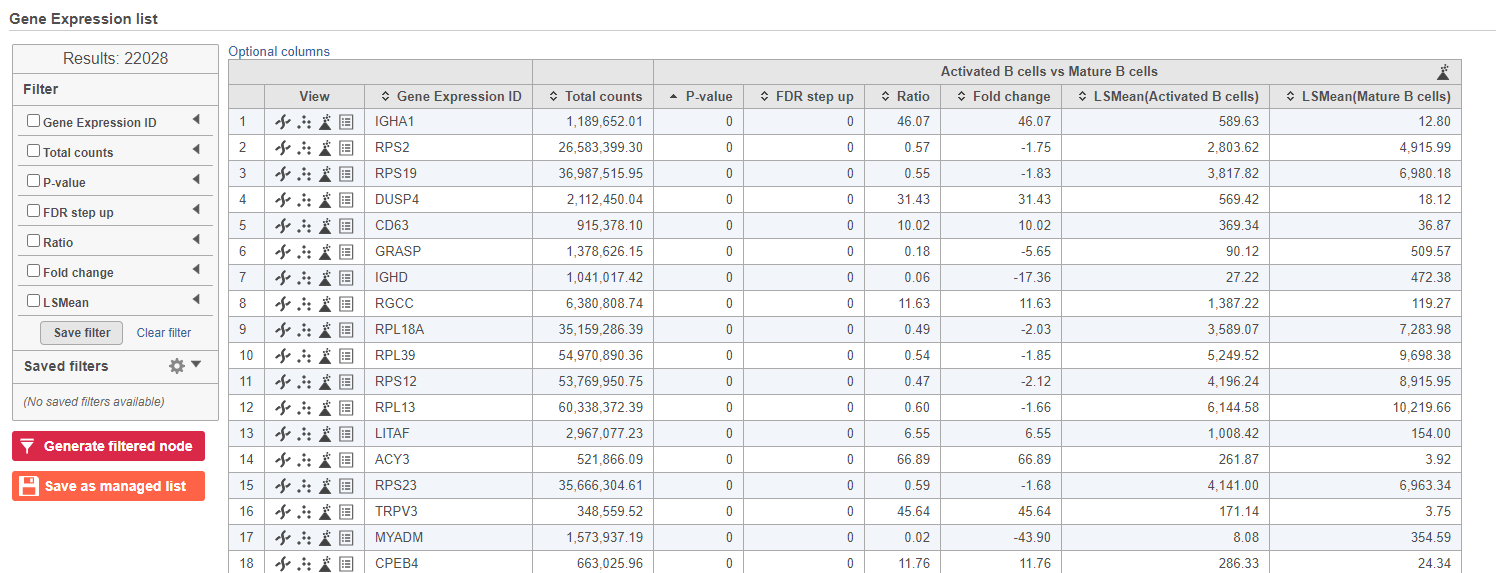 Image Added Image Added
|
Because more than 20,000 genes have been analyzed, it is useful to use a volcano plot to get an idea about the overall changes.
- Click
 Image Added in the top right corner of the table to open a volcano plot
Image Added in the top right corner of the table to open a volcano plot
The Volcano plot opens in a new data viewer session, in a new tab in the web browser. It shows each gene as a point with cutoff lines set for P-value (y-axis) and fold-change (x-axis). By default, the P-value cutoff is set to 0.05 and the fold-change cutoff is set at |2| (Figure ?).
The plot can be configured using various options in the Configuration card on the left. For example, the Color, Size and Shape cards can be used to change the appearance of the points. The X and Y-axes can be changed in the Data card. The Significance card can be used to set different Fold-change and P-value thresholds for coloring up/down-regulated genes.
| Numbered figure captions |
|---|
| SubtitleText | The volcano plot can be configured using various options in the Configuration and Selection cards |
|---|
| AnchorName | Volcano plot gene expression |
|---|
|
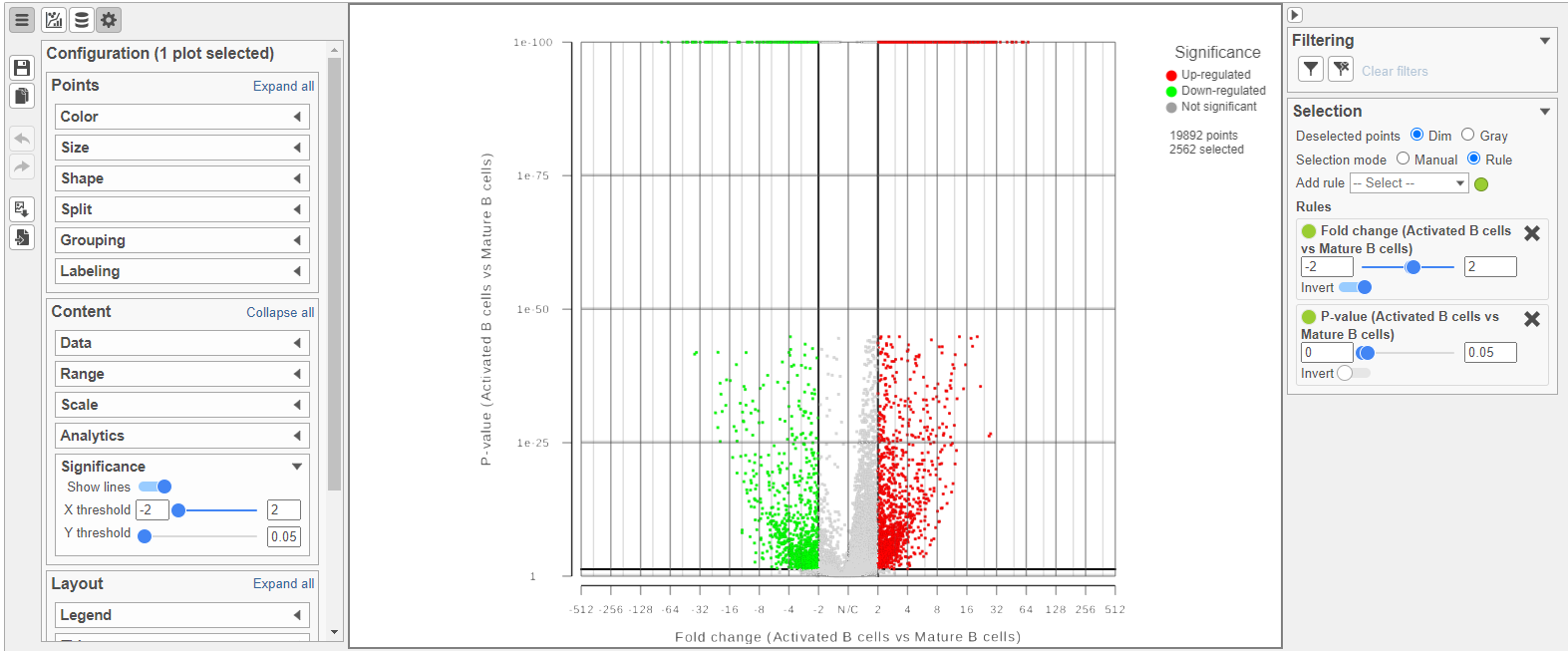 Image Added Image Added
|
- Click the GSA report tab in your web browser to return to the full report
We can filter the full set of genes to include only the significantly different genes using the filter panel on the left.
- Click FDR step up
- Type 0.05 for the cutoff and press Enter on your keyboard
- Click Fold change
- Set to From -2 to 2 and press Enter on your keyboard
The number at the top of the filter will update to show the number of included genes (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | Use the panel on the left to filter the list for significant genes |
|---|
| AnchorName | Significant genes |
|---|
|
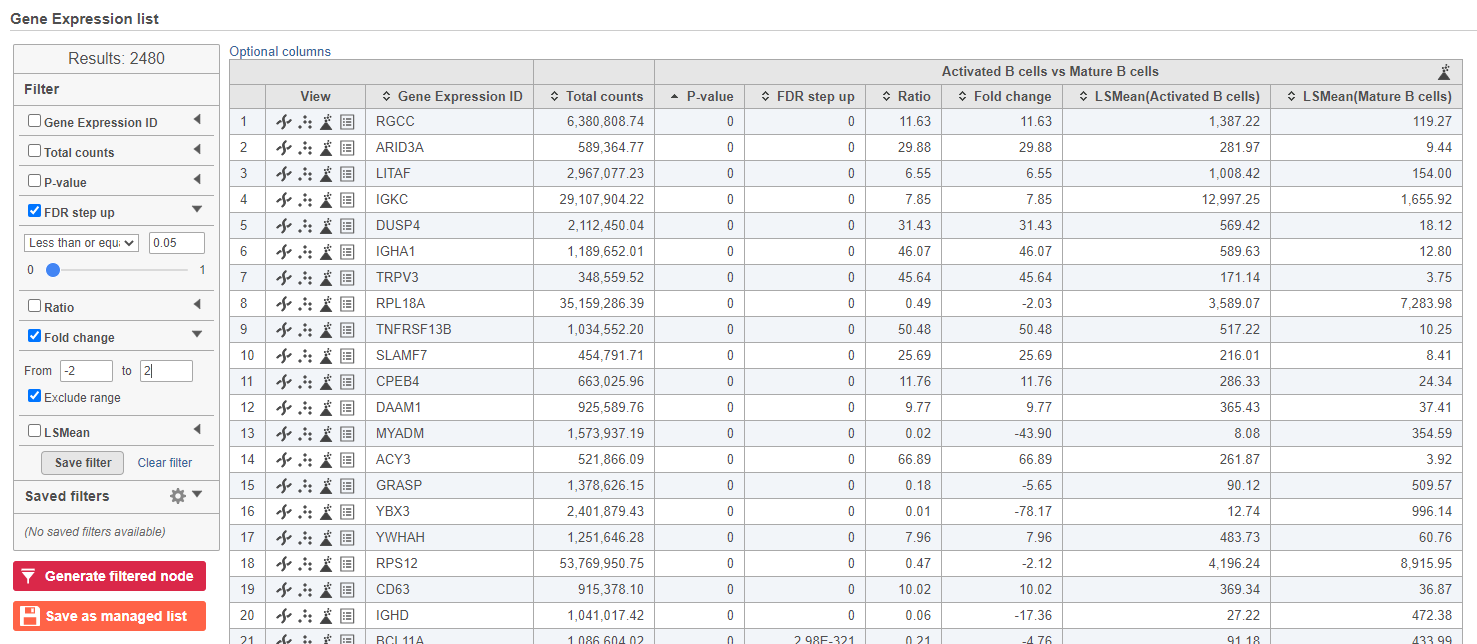 Image Added Image Added
|
- Click
 Image Added to create a new data node including only these significantly different genes
Image Added to create a new data node including only these significantly different genes
A task, Differential analysis filter, will run and generate a new Filtered Feature list data node. We can get a better idea about the biology underlying these gene expression changes using gene set or pathway enrichment. Note, you need to have the Pathway toolkit enabled to perform the next steps.
- Click the Filtered feature list data node
- Click Biological interpretation in the toolbox
- Click Pathway enrichment
- Make sure that Homo sapiens is selected in the Species drop-down menu
- Click Finish to run
- Double-click the Pathway enrichment task node to open the task report
The pathway enrichment results list KEGG pathways, giving an enrichment score and p-value for each (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | Results of pathway enrichment test |
|---|
| AnchorName | Pathway enrichment analysis results |
|---|
|
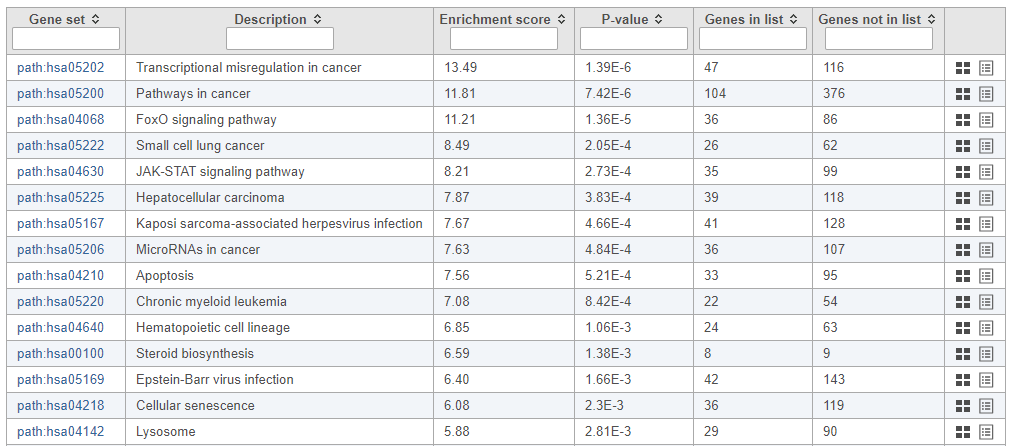 Image Added Image Added
|
To get a better idea about the changes in each enriched pathway, we can view an interactive KEGG pathway map.
- Click path:hsa05202 in the Transcriptional misregulation in cancer row
The KEGG pathway map shows up-regulated genes from the input list in red and down-regulated genes from the input list in green (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | Transcriptional misregulation in cancer pathway with significant genes highlighted in green and red |
|---|
| AnchorName | Transcriptional misregulation in cancer |
|---|
|
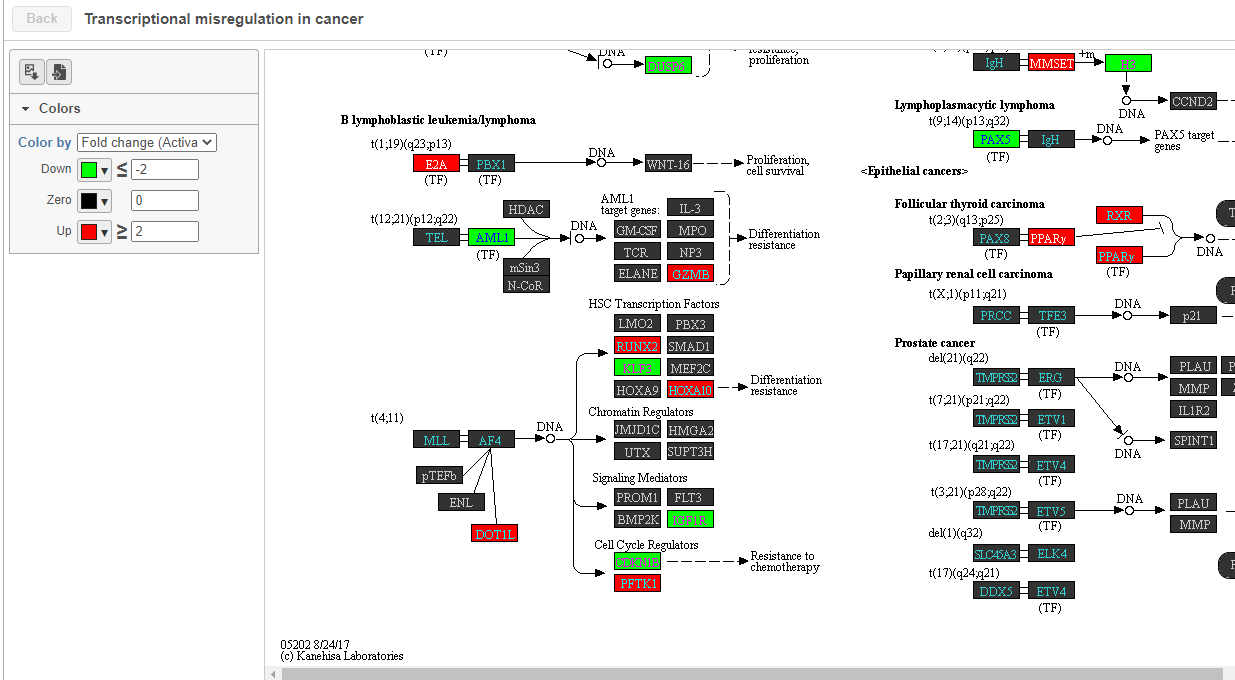 Image Added Image Added
|
| Numbered figure captions |
|---|
| SubtitleText | Final CITE-Seq pipeline |
|---|
| AnchorName | CITE-Seq final pipeline |
|---|
|
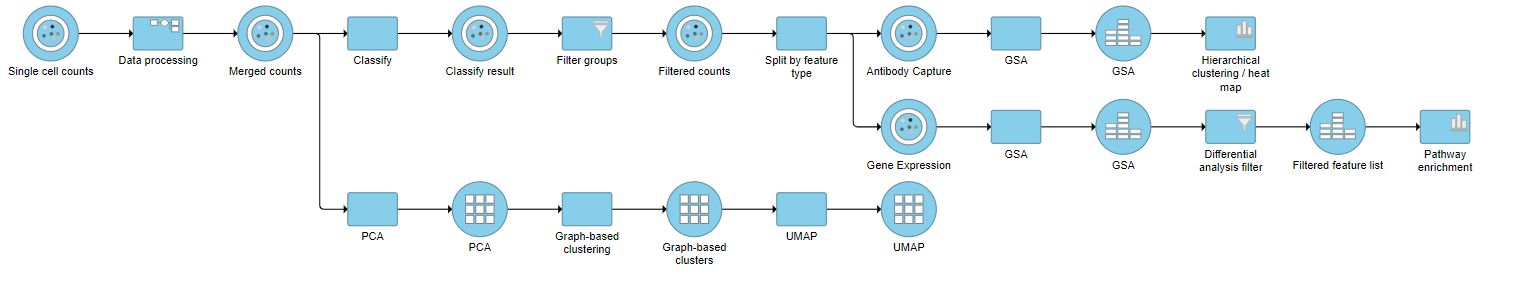 Image Added Image Added
|
...










