...
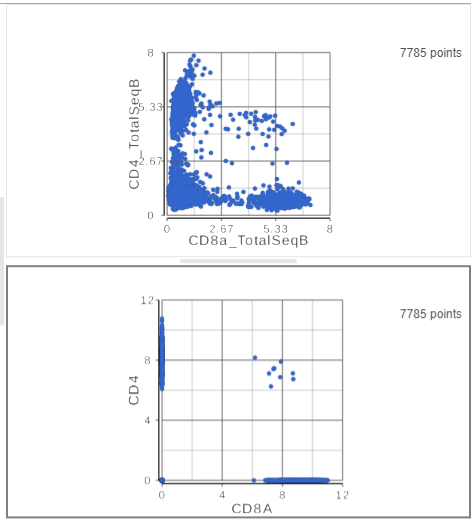
The second 2D scatter plot has the CD8A and CD4 mRNA markers on the x- and y-axis, respectively (Figure ?). The protein expression data has a better dynamic range than the gene expression data, making it easier to identify sub-population. This an advantage of CITE-Seq data over single cell RNA-Seq data alone.
| Numbered figure captions |
|---|
| SubtitleText | The second 2D scatter plot (bottom) has the CD8 and CD4 genes plotted against each other |
|---|
| AnchorName | 2nd 2D scatter plot |
|---|
|
|
- On the first 2D scatter plot (with protein markers), click
 Image Added in the top right corner
Image Added in the top right corner - Manually select the cells with high expression of the CD4_TotalSeqB protein marker (Figure ?)
More than 2000 cells show positive expression for the CD4 cell surface protein.
| Numbered figure captions |
|---|
| SubtitleText | Draw a lasso to manually select CD4+ cells, based on protein expression |
|---|
| AnchorName | Select CD4+ cells (protein) |
|---|
|
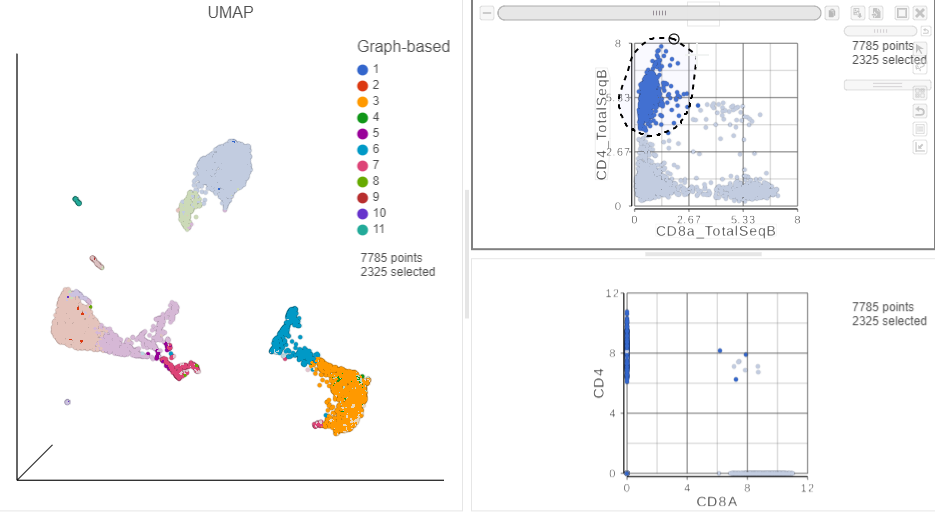 Image Added Image Added
|
- Click
 Image Added in the top right of the plot to switch back to pointer mode
Image Added in the top right of the plot to switch back to pointer mode - Click on a blank spot on the plot
- On the second 2D scatter plot (with mRNA markers), click
 in the top right corner
in the top right corner - Manually select the cells with high expression of the CD4 gene marker (Figure ?)
...
| Numbered figure captions |
|---|
| SubtitleText | Draw a lasso to manually select CD4+ (mRNA) cells |
|---|
| AnchorName | Select CD4+ cells |
|---|
|
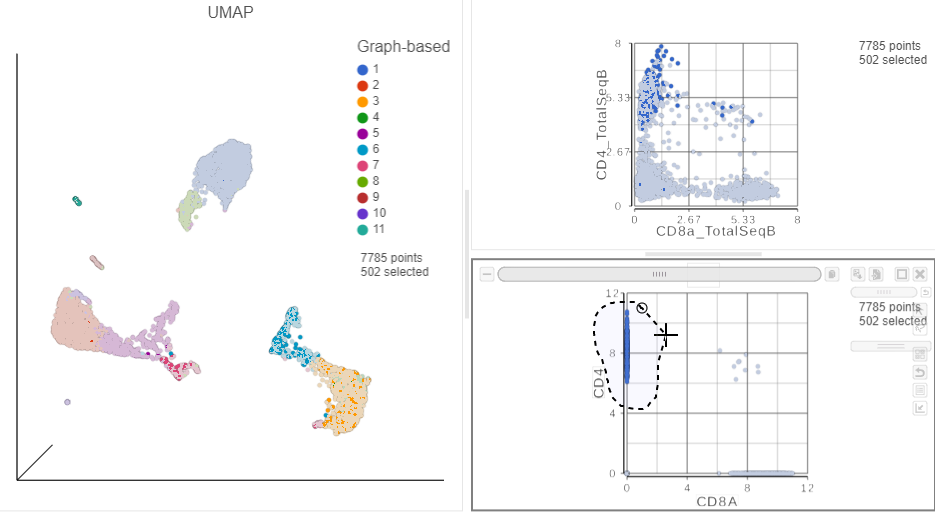 Image Added Image Added
|
This time, approximately 500 cells show positive expression for the CD4 marker gene. This means that the protein data is also less sparse than the gene expression data.
T cells
Based on the exploratory analysis above, most of the CD3 positive cells are in the group of cells in the bottom right corner of the UMAP plot. This is likely to be a group of T cells. We will now examine this group in more detail to identify T cell sub-populations.
- Click
 Image Added in the top right corner of both 2D scatter plots, to remove them from the canvas
Image Added in the top right corner of both 2D scatter plots, to remove them from the canvas - Click
 Image Added in the top right corner of the 3D UMAP plot
Image Added in the top right corner of the 3D UMAP plot - Draw a lasso around the group of putative T cells (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Select the group of putative T cells |
|---|
| AnchorName | Lasso T cells |
|---|
|
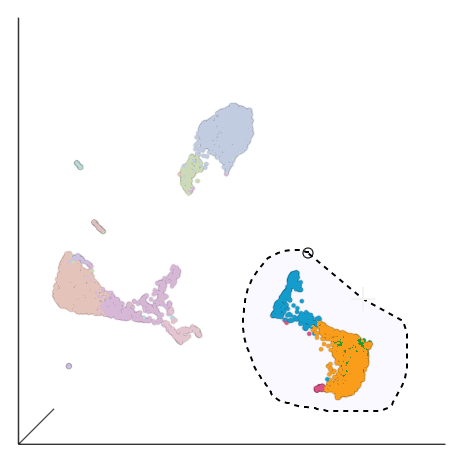 Image Added Image Added
|
- Click
 Image Added to include the selected points
Image Added to include the selected points - Click
 Image Added in the top right of the plot to switch back to pointer mode
Image Added in the top right of the plot to switch back to pointer mode - Click and drag the plot to rotate it around
Deselected cells are excluded and the axes have been rescaled to give better resolution of the selected points (Figure ?). Note that the UMAP has not been recalculated, the axes have just been rescaled.
| Numbered figure captions |
|---|
| SubtitleText | Group of putative T-cells |
|---|
| AnchorName | T cell group |
|---|
|
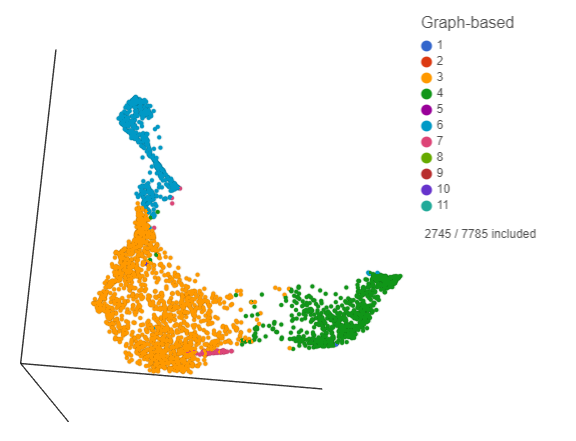 Image Added Image Added
|
This group of putative T cells predominantly consists of cells assigned to graph-based clusters 3, 4, 6, and 7. Examining the biomarker table for these clusters can help us infer cell types.
- Click and drag the bar between the UMAP plot and the biomarker table to resize the biomarker table to see more of it (Figure ?)
If you need to create more space on the canvas, hide the selection panel using the  Image Addedicon on the right and/or the
Image Addedicon on the right and/or the  Image Added icon to hide the menu on the left.
Image Added icon to hide the menu on the left.
| Numbered figure captions |
|---|
| SubtitleText | Resize plots to see more of the biomarker table |
|---|
| AnchorName | CITE-Seq biomarker table |
|---|
|
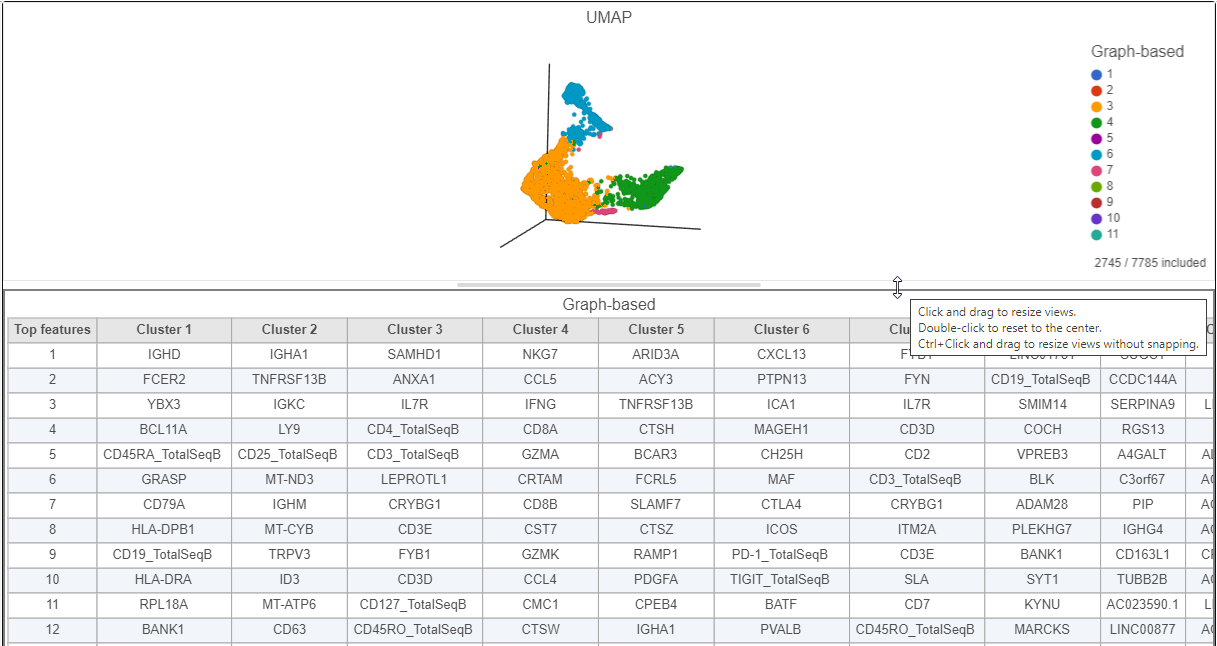 Image Added Image Added
|
B cells
...





