Page History
...
- Click the Single cell counts data node
- Click Pre-analysis tools in the toolbox
- Click Split by feature type
A rectangle, or rectangular task node , will be created for Split matrix along with two output circles, or circular data nodes, one for each data type (Figure 2?). The labels for these data types are determined by features.csv file used when processing the data with Cell Ranger. Here, our data is labeled Gene Expression, for the mRNA data, and Antibody Capture, for the protein data.
...
An important step in analyzing single cell RNA-Seq data is to filter out low-quality cells. A few examples of low-quality cells are doublets, cells damaged during cell isolation, or cells with too few coutns counts to be analyzed. In a CITE-Seq experiment, protein aggregation in the antibody staining reagents can cause a cell to have a very high number of counts. These are low-quality cells that can be excluded. Additionally, if all cells in a data set are expected to show a baseline level of expression for one of the antibodies used, it may be appropriate to filter out cells with very low counts or a low number of detected features. You can do this in Partek Flow using the Single cell QA/QC task.
...
- Double-click the Single cell QA/QC task node to open the task report
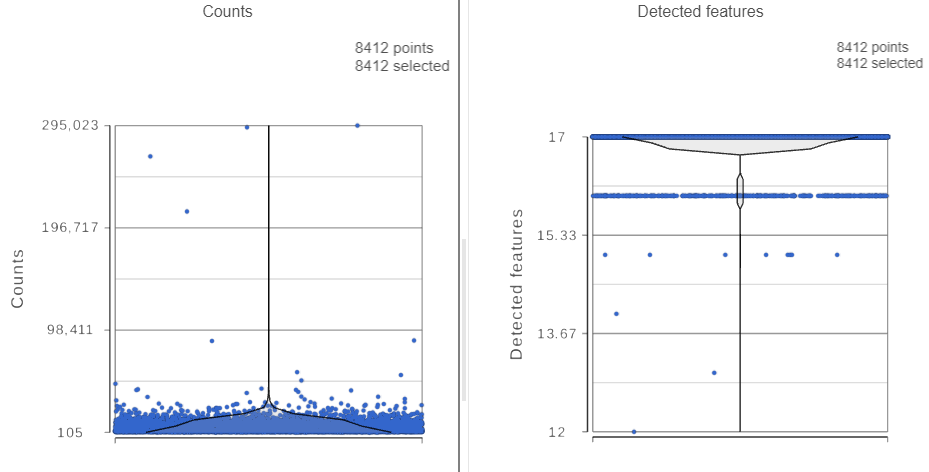
The task report lists the number of counts per cell and the number of detected features per cell in two violin plots. For more information, please see our documentation for the Single cell QA/QC task. For this analysis, we will set a maximum counts threshold to exclude potential protein aggregates and, because we expect every cell to be bound by several antibodies, we will also set a minimum counts threshold.
- Click the Single cell QA/QC node once it finishes running
- Click Task report on the task menu
The Single cell QA/QC report opens in a new data viewer session. There are interactive violin plots showing the most commonly used quality metrics for each cell from all samples combined (Figure ?). For this data set, there are two relevant plots: the total count per cell and the number of detected genes features per cell. Each point on the plots is a cell and the violins illustrate the distribution of values for the y-axis metric. Because mitochondrial transcripts are not present in protein data, this plot is not informative for this data set.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
For this analysis, we will set a maximum counts threshold to exclude potential protein aggregates and, because we expect every cell to be bound by several antibodies, we will also set a minimum counts threshold.
- Select one of the plots on the canvas
- In the Selection card on the right, set the Counts threshold to keep cells between 500 and 20000 (Figure ?)
...
The recommended normalization for protein data includes the following steps: Add 1, Divide by Geometric mean, Add 1, log Log base 2. This is a variant of Centered log-ratio (CLR), which was used to normalize antibody capture protein counts data in the paper that introduced CITE-Seq [1] and in subsequent publications on similar assays [2. 3]. CLR normalization includes the following steps: Add 1, Divide by Geometric mean, Add 1, log base e. Normalizing the protein data to base 2 instead of e allows for better integration with gene expression data further downstream. If you would prefer to use CLR, click and drag CLR from the panel on the left to the right.
...
Next, we can normalize the mRNA data. We will use the recommended normalization method in Partek Flow, which accounts for differences in library size, or the total number of UMI counts, per cell, adds 1 and log log2 transforms the data.
- Click the Filtered counts data node produced by filtering the Gene Expression data node
- Click the Normalization and scaling section in the toolbox
- Click Normalization
- Click the button
- Click Finish to run (Figure ?)
...
- Double-click Data processing to expand it , or right-click and choose Expand collapsed task
...