Page History
...
Partek distributes prep kits for processing several types of data - Drop-seq, 10x Chromium Single Cell 3' v2, Fluidigm C1 mRNA Seq HT IFC, and Rubicon Genomics ThruPLEX Tag-seq. If your data is from one of these sources, you can select the appropriate option in the Prep kit drop-down menu. If the data is from another source, you can build a custom prep kit file to process your data.
...
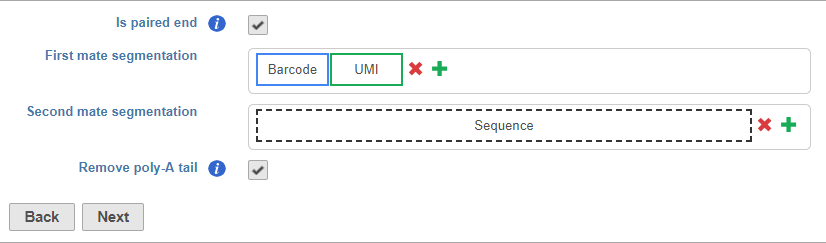
Segments include adaptors, barcodes, UMIs, and the insert (i.e., the target sequence of the assay - mRNA in RNA-Seq)
Adaptors
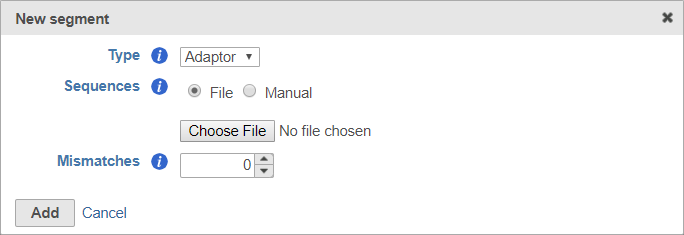
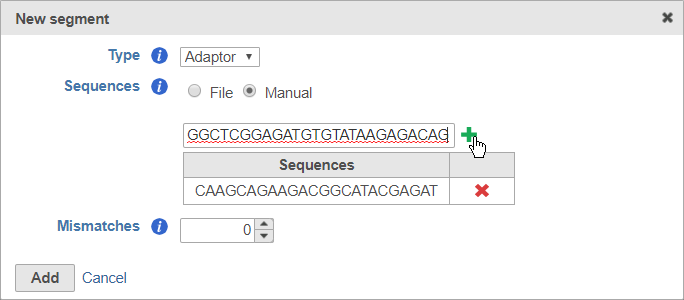
For adaptors, you have the option of choosing a file with your adaptor sequences or entering the adaptor sequences manually.
To use a file, choose File for Sequences and then click Choose File (Figure 6). Use the file browser to choose a FASTA file from your local computer.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
You can choose to specify the mismatch allowance using the Mismatches option.
After you have specified the file or manually entered the sequences, click Add to add the adaptor sequence(s).
UMIs
Unique Molecular Identifiers (UMIs) are randomly generated sequences that uniquely identify an original starting molecule after PCR amplification.
Including a UMI in your prep kit will allow you to access a downstream tasks task that uses UMI information, Deduplicate UMIs. For more information about Deduplicate UMIs, please see our UMI Deduplication in Partek Flow white paper. While the UMI sequence will be trimmed, a record of the UMI sequence for each read is retained for use by downstream tasks.
...
Like adaptors, barcodes can be specified using a file or manually specified, but you can also choose to designate any a segment of arbitrary length in the sequence as the barcode. This is useful if you do not have a specific set of known barcode, but do have a segment in your sequence that servers as the barcodebarcodes.
To set the barcode to an arbitrary segment, choose Arbitrary and specify the barcode length (Figure 9).
...
Remember to click Add to add the new segment to your prep kit.
Insert
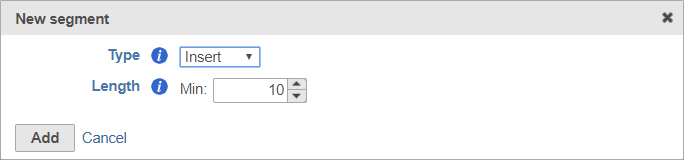
The insert is the target of your sequencingsequence retained after trimming in the Trimmed reads data node. For example, in RNA-Seq, this would be the mRNA sequence. Every prep kit must include an insert segment. You can specify the minimum size of the insert section using the Length field (Figure 10). The insert segment is the part of each read (or pair of reads) that is retained in the Trimmed reads output data node.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Prep kits download as a .zip file. This Prep kit .zip file can be added to imported into Partek Flow by selecting Import from a file when adding a new prep kit. Select the .zip file when importing, do not unzip the file.
...