Page History
...
This tutorial will illustrate how to:
| Pgs7 note |
|---|
|
Introduction to Survival Analysis
...
An important feature of survival analysis is the presence of “censored” data. For example, medical studies often focus on survival of patients after treatment so the survival times are recorded during the study period. At the end of the study period, some patients are still alive, some have died (and survival data should be available for these)dead, some patients are alive, and the status of some patients is unknown because they dropped out of the study. Censored data represent the last group (study drop-outs). The information from censored data is valuable because while it does not measure the actual survival time, it does provide information about survival up until the patient refers to the latter two groups. Neither the patients who survived until the end of the study or those who dropped out of the study . Within the field of survival analysis, special tests are developed to correctly use both censored and uncensored observations. Details about the statistical tests implemented in Partek® Genomics Suite™ software can be found in the user's manual. have experienced the study event, "death".
Tutorial Data Set
This example data set (236 samples) is a subset of fresh-frozen breast tumor specimens from a population-based cohort of 315 women with breast cancer. The clinicopathological characteristics accompanying each tumor include p53 status (mutant or wild-type), estrogen receptor (ER) status, progesterone receptor (PgR) status, lymph node status, tumor size, and patient age. Gene expression of all the samples was assessed on Affymetrix® U133A and U133B arrays (Miller LD et al., GSE3494). Please note that Affymetrix data have been chosen for the illustration purposes only, and that the same functionality can be used to analyze data generated by any vendor. The raw data files (.CEL) have already been imported into PGS; samples with no survival time data as well as sample attributes irrelevant for the survival analysis were removed, and the final spreadsheet was saved in Partek Genomics Suite (Survival_Tutorial.fmt and Survival_Tutorial.txt). To download the tutorial data set, use this link - Survival Tutorial Data. Unzip the downloaded folder and save it in an easily accessible location on your computer.
After saving the unzipped file, you can open it in Partek Genomics Suite.
- Select File from the main toolbar
- Select Open...
- Browse to the folder containing the tutorial data set and select the file Survival_Tutorial.fmt
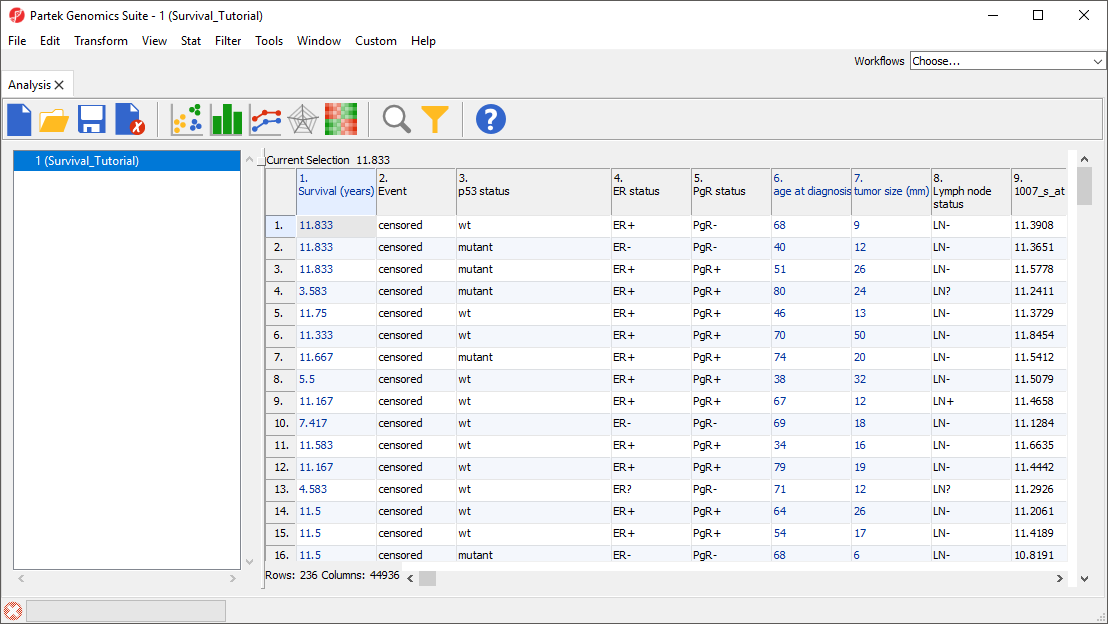
The data spreadsheet will open (Figure 1). Each row represents a tumor sample from a breast cancer patient. Sample attributes are listed in columns 1-8, while columns 9+ are intensity values for the probe sets listed in the column headers.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
...