Page History
...
For Partek® Genomics Suite® to recognize an annotation spreadsheet, it must meet several requirements. First, there must be a column header row in the annotation file. Second, there must be a column in the annotation file that matches the probe identifiers in your data spreadsheet. Third, any text field above the column header row must start with #. Fourth, the text fields must be tab or comma delimited.
...
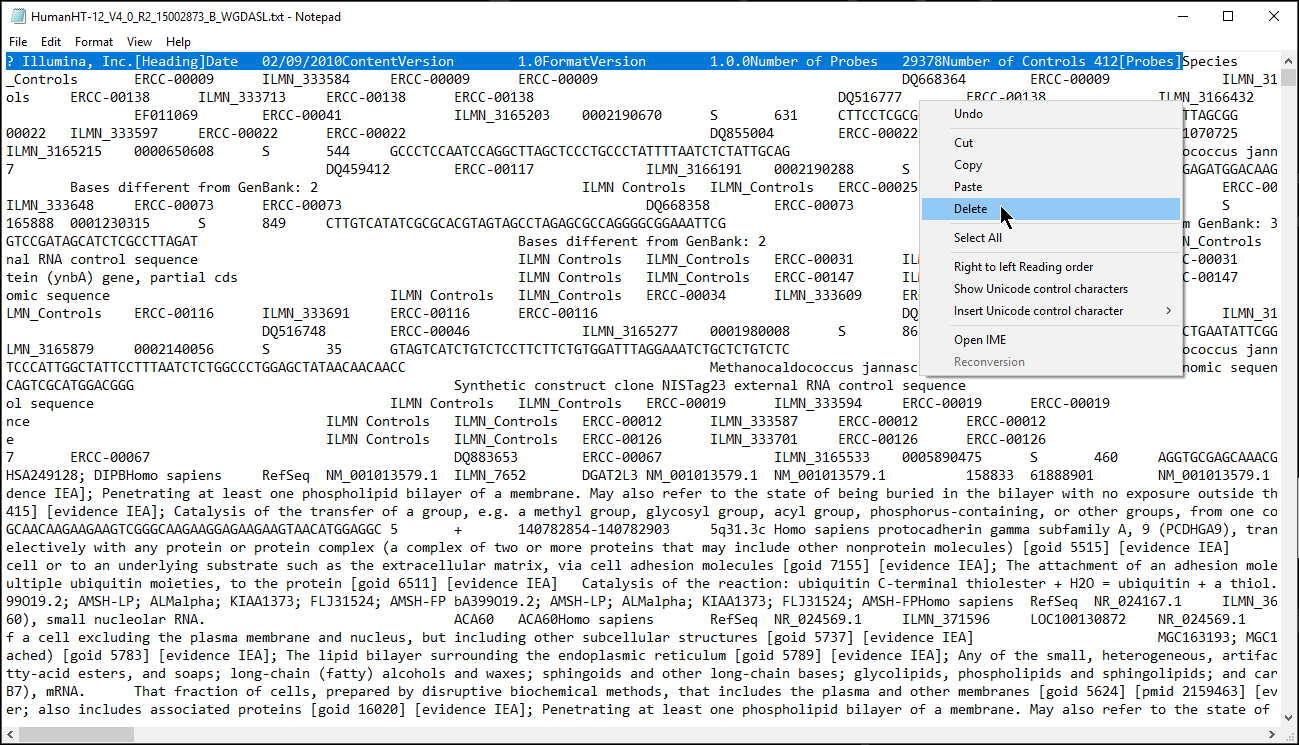
- Open the annotation file with a text editor such as Notepad++/WordPad/TextEdit (Microsoft Excel is not recommended to edit text files, for instance when used default settings, it converts gene names to dates and floating-point numbers)
- Verify that a column in the annotation file matches the probe identifiers identifier in your data spreadsheet, e.g probe ID, the identifier has to be unique
- Remove the text before the first column header (Figure 1) or add # to each text box and save the annotation file
If it is difficult to visualize where the column headers start, it might help to import the annotation file into a spreadsheet viewing program such as Microsoft Excel or Google Sheets.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
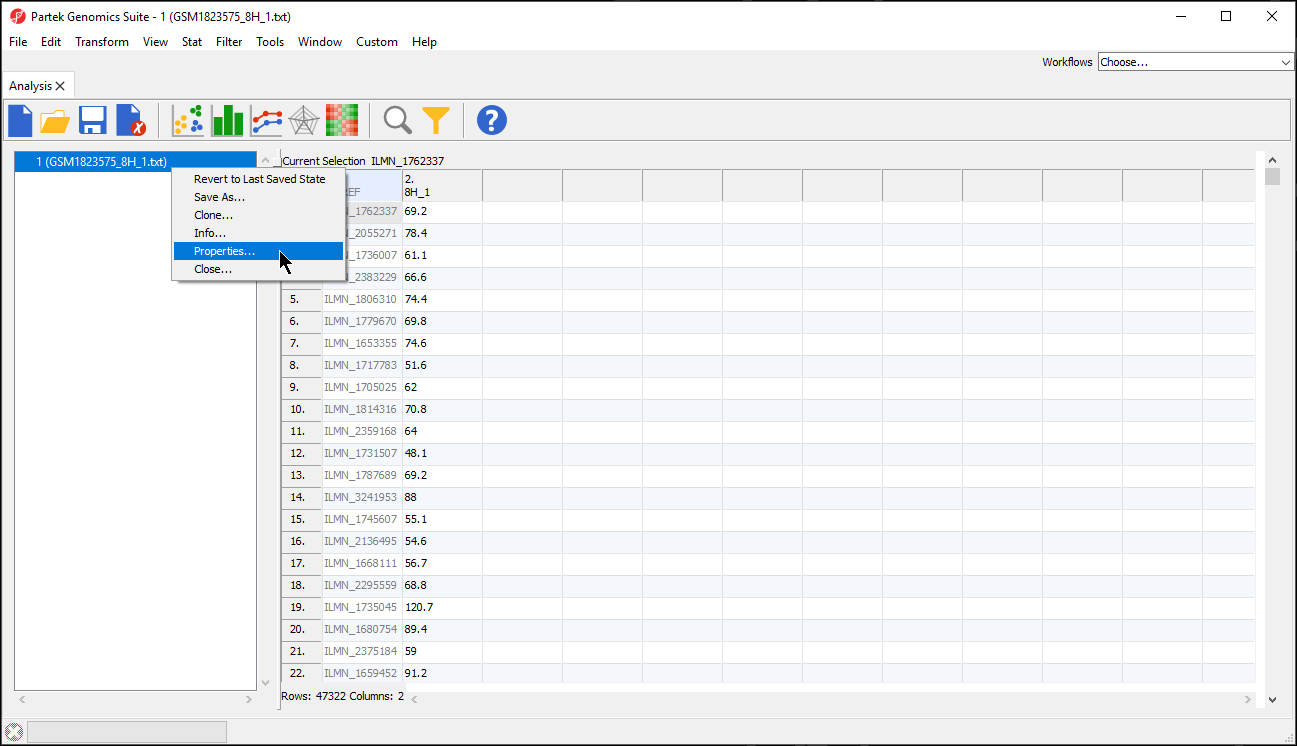
- Right-click the spreadsheet you want to annotate in the spreadsheet tree panelSelect , select Properties from the pop-up menu (Figure 2) or
- Choose File > Properties from the menu on the selected spreadsheet
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
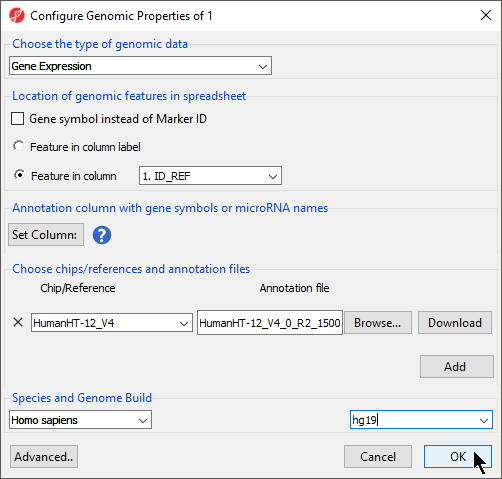
The Configure Genomic Properties dialog will now open. Select the appropriate option for Choose the type of genomic data; here we have chosen Gene Expression. Expression (Figure 4).
Location of genomic features in spreadsheet gives option to specify whether genomic features (e.g. genes, miRNAs, probes, SNPs, CpGs etc) are represented by columns or rows:
- Feature in column label: each feature is on a column, each row is a sample
- Feature in column: each feature is on a row, the feature ID is a specified column number
- When Gene Symbol instead of Marker ID is checked, annotation file is not needed when perform biological interpretation, since PGS can use the gene symbol to look up gene set/pathway database
Choose chips/reference and annotation files section allow you to specify feature annotation file associates to the current spreadsheet
- Select Browse... from Choose chips/references and annotation files
- Select your annotation spreadsheet file using the file selection interface
...
- Select Close to return to the Configure Genomic Properties
An index file is generated in the same folder as the annotation file, it has the same file name as the annotation file, but the extension name is .idx. If you need to re-configure the genomic location field in the annotation file, you need to manually delete the .idx file and re-do the above step to re-generate .idx for the annotation file.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The Chip/Reference text field will be populated with the annotation file name. You can edit this text field this if you wish.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Annotation column with gene symbols or miRNA names: in the annotation file, if the gene symbol field is labeled as Gene Symbol, PGS is using this field to look up gene set/pathway database, however, if the gene symbol/or miRNA name field is labels as something else, e.g. SystematicName, you need to manually specify this field as gene symbol field.
- Click on Set Column: button to view/ or select the field corresponding gene symbol information
Note: Species and gene symbol information is required for biological interpretation analysis.
- Select OK apply the annotation file to your data spreadsheet
To verify that the annotation has been added, we can try to add annotation information to the spreadsheet when the feature are on rows in the spreadsheet.
- Right-click on a column in the annotated data file spreadsheet
- Select Insert Annotation from the pop-up menu (Figure 5)
...
Annotation files for most commercial arrays are available from the chip manufacturer. If you have a custom chip or want to use a customized annotation file, you can create an annotation file that will allow you to add annotations to your data spreadsheet and invoke a Probe Set HTML reportfeatures (e.g. probe IDs) when the features are represented by rows on the spreadsheet. Your annotation file must meet the following criteria:
...
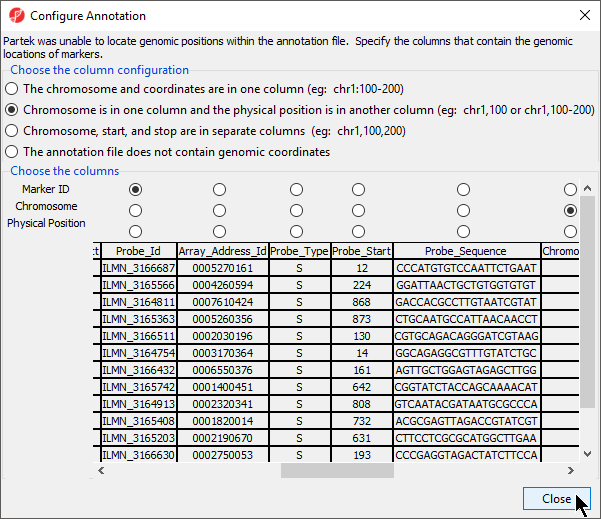
The annotation file must also contain a column that has the chromosome and base pair location (start and stop or physical position). Cytoband and/or strand can also be included. The table below provides possible column labels, a description of the format for that field, and an example.
Note: In this table, the examples are for a gene on the top strand of chromosome 3; on the p arm in cytoband 14.2 starting at 69,871,322 base pairs and ending at 70,100,176
| Column label | Description of format | Example |
|---|---|---|
| chromosome | a chromosome label | 3 |
| start | an integer, the start position (in base pairs) of the feature | 69871322 |
| stop | an integer, the stop position (in base pairs) of the feature | 70100176 |
| genomic_coordinates | chromosome:start-stop | 3:69871322-70100176 |
| strand | + for top, - for bottom | + |
| physical position | an integer, the position (in base pairs) of the feature | 70100176 |
...