Page History
...
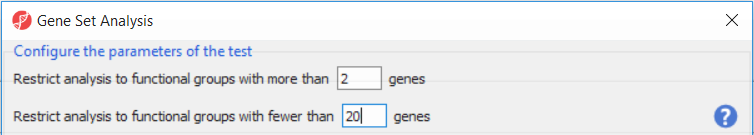
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
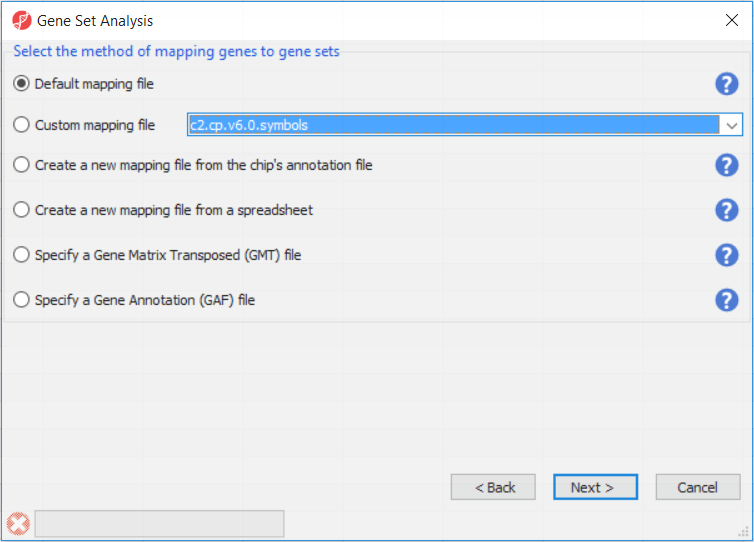
The next dialog (Figure 2) specifies the method of mapping genes to gene sets. Default mapping file is built from annotation files from geneontology.org. Custom mapping file points to the mapping files available on the local computer and present in the Microarray libraries directory. Create a new mapping file from the chip's annotation file option will try to build the annotation file from the annotation file created by the microarray vendor. Create a new mapping file from a spreadsheet enables you to create a custom mapping file from an open spreadsheet, which has gene symbols on one column, and gene groups on the other column. Finally, files in gene matrix transposed (GMT) or gene annotation (GA) formats can also be used.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|
Overview
Content Tools