Page History
...
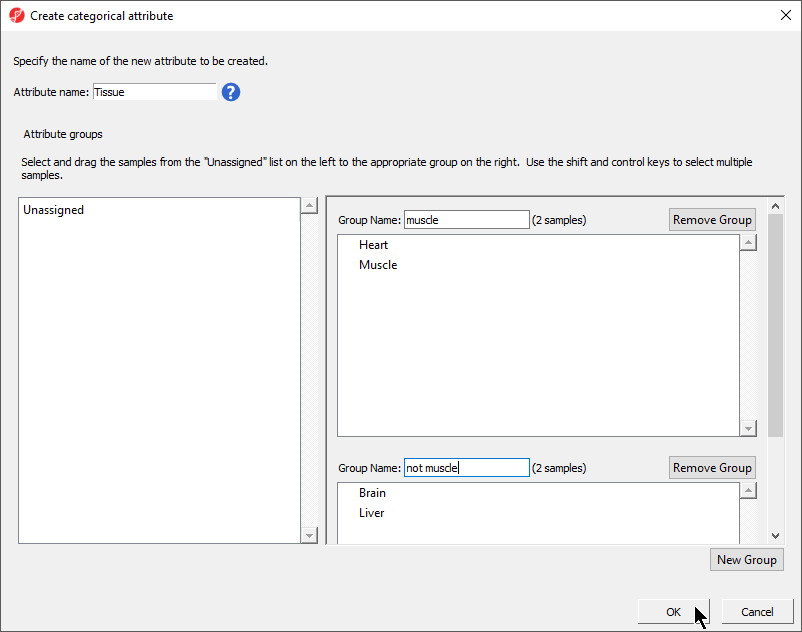
Creating a categorical sample attribute allows us to group samples. This is useful for designating samples as replicates, as members of an experimental group, or as sharing a phenotype of interest. In this tutorial, we have four different samples from different tissues and different donors, but to illustrate the available statistical analysis options later on, we need to group the samples into two groups: muscle (heart Heart and muscleMuscle) and not muscle (brain Brain and liverLiver).
- Set Attribute name: as Tissue
- Rename Group 1 to muscle and Group 2 to not muscle
- Select and drag the samples from the Unassigned panel to the correct group panel (Figure 5)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select OK
- Select No from the Would you like to create Add another categorical attribute? dialog
- Select Yes from theWould you like to save the spreadsheet the Save spreadsheet 1 dialog
The attribute will now appear as a new column in the RNA-seq spreadsheet with the heading Tissue and the groups muscle and not muscle.
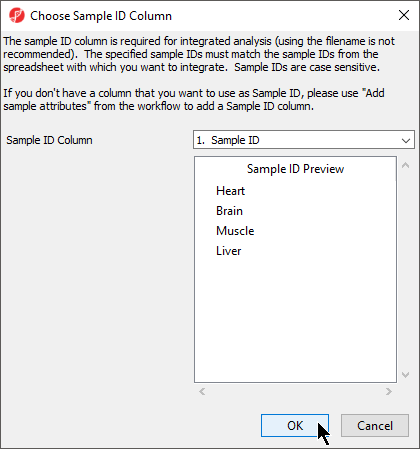
The next available step in the Import panel of the RNA-seq workflow is Choose Sample ID Column. Verifying the correct column is designated the Sample ID becomes particularly important when data from multiple experiments is being combined.
- Select Choose Sample ID Column from the Import panel of the RNA-seq workflow
- Select OK (Figure 6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Rate Macro | ||
|---|---|---|
|