The original experiment is listed on the Gene Expression Omnibus as GSE848; however, this tutorial only uses a subset of the original experiment and should be downloaded from the Partek website tutorial page, Gene Expression Analysis with Batch Effects.
- Download the zipped project folder, Breast_Cancer-GE.zip
- Unzip the project folder to C:/Partek Training Data/ or a directory of your choosing
This location should be easily accessible. The unzipped Breast_Cancer-GE project folder and a zipped annotation file will be added to the selected directory.
- Unzip the included annotation file, HG_U95Av2.na32.annot.rar
- Move the annotation file, HG_U95Av2.na32.annot, to the microarray libraries folder
By default, the microarray libraries folder will be located at C:/Microarray Libraries, but the location may vary depending on your operating system and configuration.
- Open Partek Genomics Suite
- Select () from the main command bar
- Navigate to the tutorial folder, Breast_Cancer-GE
- Select Breast_Cancer.txt
- Select Open (Figure 1)
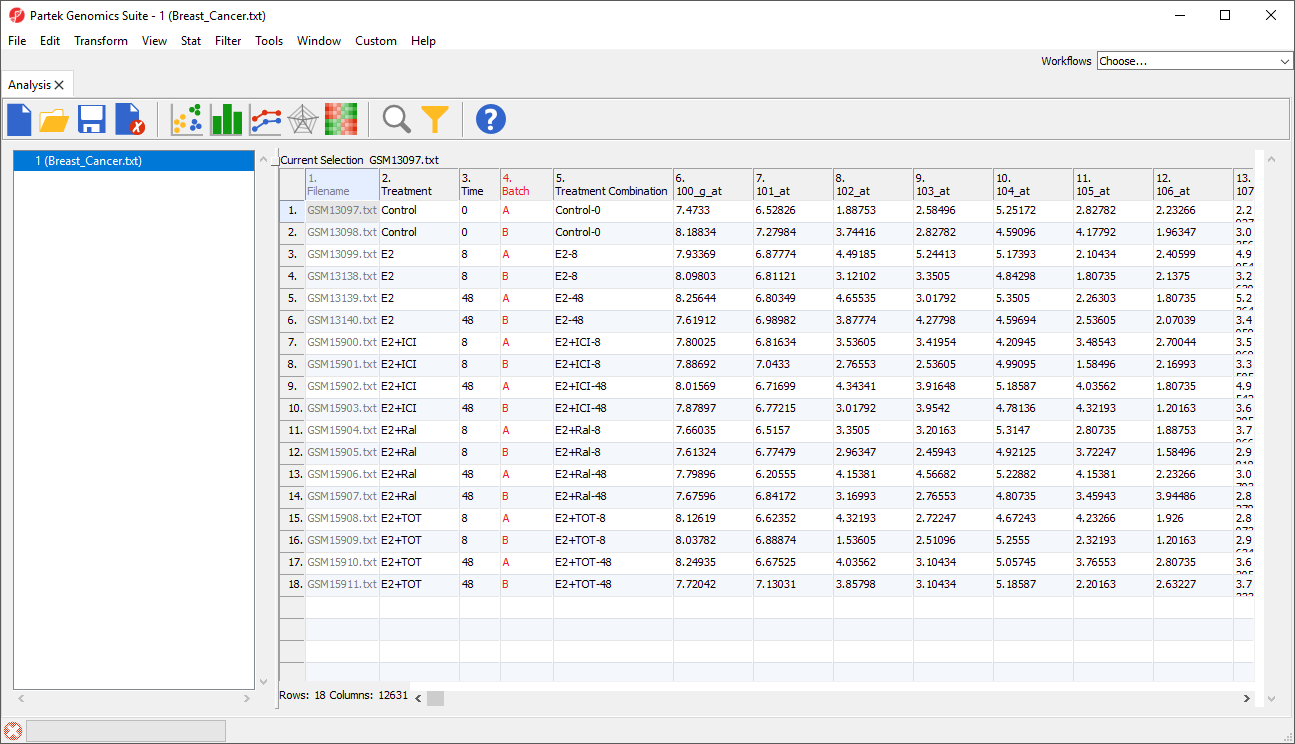
The spreadsheet will open as 1 (Breast_Cancer.txt) (Figure 2).
The summary at the bottom the spreadsheet shows there are 18 rows and 12,631 columns in the spreadsheet. The first column contains the Filename listing the GEO GSM number. This is also is an identifier for the microarray. Treatment, Time, and Batch are in columns 2, 3, and 4, respectively. Column 6 marks the beginning of the probesets. The data is log2 transformed.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
44 | rates |