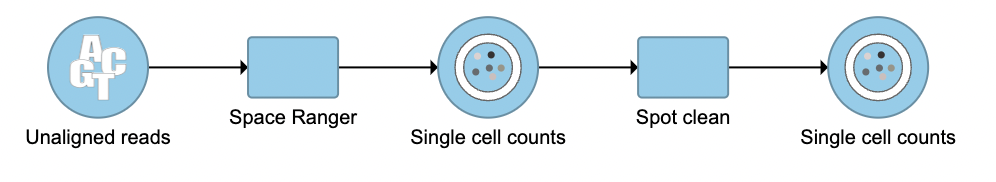
The terminology spot swapping describes the artifact for spatial data that mRNA bleed from nearby spots causes substantial contamination of UMI counts[1]. Spot clean in Flow is a task that aims to improve estimates of expression by correcting for spot swapping.
The task can only be invoked from the Space Ranger task output data node since it takes the raw count matrix as input. To run the Spot clean task in Flow:
- Click the Single cell counts outputted from Space Ranger (Figure 1)
- Click Pre-analysis tools in the toolbox
- Click Spot clean
- Click Finish to run the task with default settings
Another single cell counts node will be generated. The data node contains a matrix of cell counts with the decontaminated gene expressions (Figure 2). Downstream analysis tasks, such as normalization, PCA, and ANOVA, can be performed on the new single cell counts node.
Parameters in this task that you can adjust include:
Gene cutoff: Filter out genes with average expressions among tissue spots below or equal to this cutoff. Default: 0.1.
Max iteration: Maximum iteration for EM parameter updates. Default: 10. Set a smaller number to save computation time.
References
Ni, Z., Prasad, A., Chen, S. et al. SpotClean adjusts for spot swapping in spatial transcriptomics data. Nat Commun 13, 2971 (2022). https://doi.org/10.1038/s41467-022-30587-y
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
7 | rates |