By following the steps in this tutorial, you have built a pipeline. You can save this pipeline for future use.
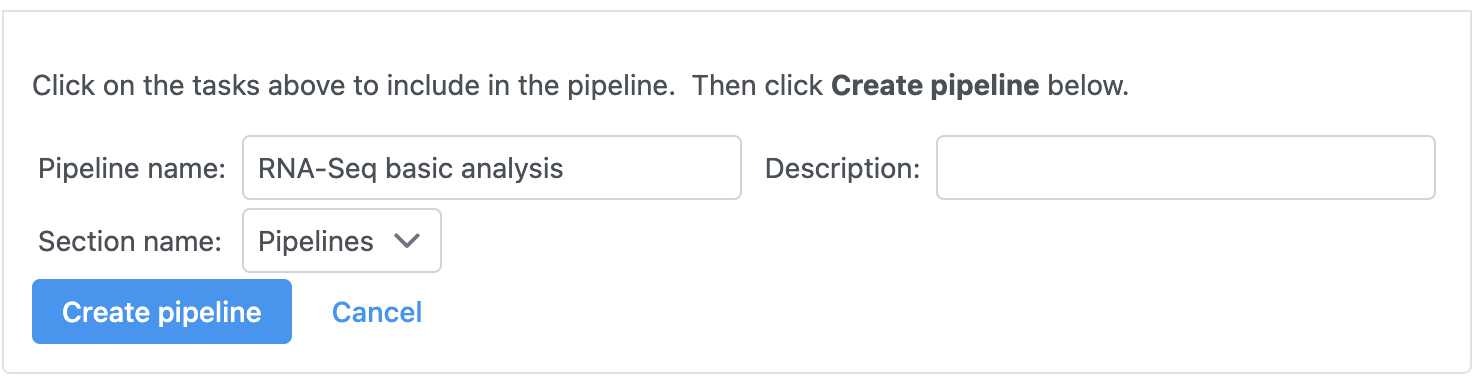
- Select Create new pipeline near the bottom left-hand side of the browser window
- Select the Pre-alignment QA/QC, Trim bases, Align reads, Post-alignment QA/QC, Quantify to annotation model, Filter features, Normalize counts, PCA, and GSA task nodes to include them in the pipeline
- Name the pipeline; we have chosen RNA-Seq basic analysis
- Give a description for the pipeline; we have noted trim <20, STAR, normalize with total count and add 0.001, GSA
- Select Create pipeline (Figure 1)
After selecting the pipeline, you will be prompted to choose the reference genome for alignment, the annotation for quantification, and the contrasts for GSA. After selecting these options, the pipeline will automatically run. See Pipelines for more information.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
32 | rates |