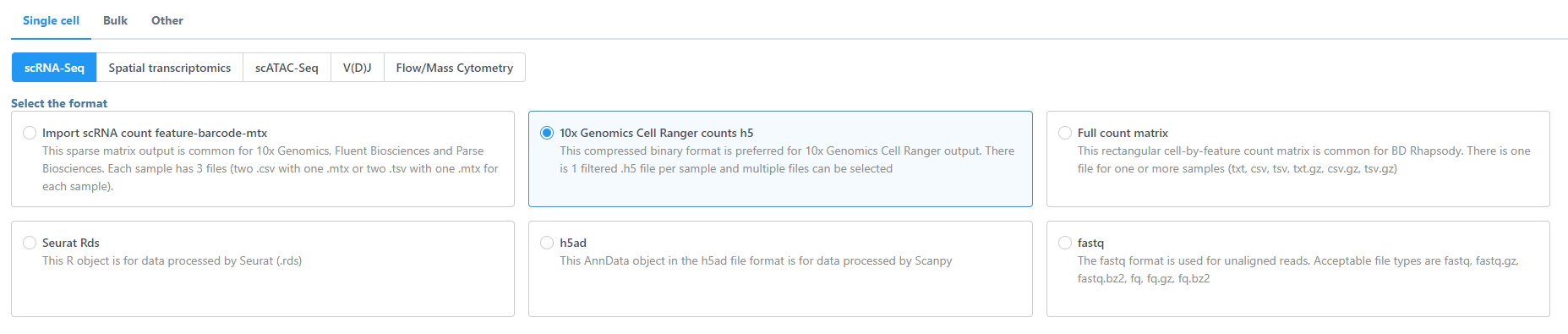
Import single cell data for different assay types and formats
Select Single cell, choose the assay type (scRNA-Seq, Spatial transcriptomics, scATAC-Seq, V(D)J, Flow/Mass cytometry), and select the data format (Figure 1). Use the Next button to proceed with import.
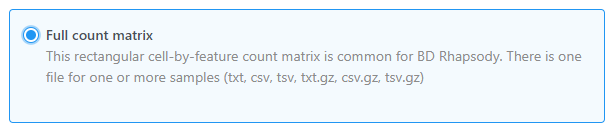
Import single cell data from count matrix text file(s) using Full count matrix as the data format
Partek Flow supports single cell data analysis in count matrix text format using the Full count matrix data format (Figure 2). Each matrix text file is assumed to represent on sample, each value in the matrix represents expression value of a feature (e.g. a gene, or a transcript) in a cell. The expression value can be raw count, or normalized count. The requirement of the format of each text file should be the same as count matrix data.
Specify text file location, only one text file (in other words one sample) can be imported at once, preview of the file will be displayed, configuration of the file format is the same as Import count matrix data. In addition, you need to specify the details about this file.
Click Finish, the sample will be imported, on the data tab, number of cells in the sample will be displayed.
To import multiple samples, repeat the above steps by clicking Import data on the Metadata tab or within the task menu (toolbox) on the Analyses tab. Make the same previous selections using the cascading menu.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
2 | rates |