Next generation sequencing (NGS) data is notably huge in file size. Dealing with NGS data is not only time consuming but also puts constraints on hard disk space. This is especially true if analysis parameters need to be optimized. The Filter reads task is a very useful tool to get a subset of the raw data upon which optimization can be performed. The optimized parameters can then be saved and applied to the whole dataset.
Filter reads is only available for unaligned reads of FASTQ format. Select the Unaligned Reads data node then select Filter reads from the Pre-alignment tools section on the menu.
There are two options to filter reads: Subsample reads and Filter by read length.
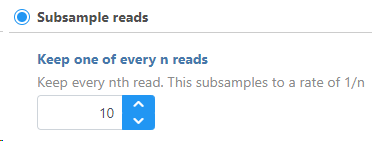
To Subsample reads, specify how many reads you want to keep for every nth reads. For example: if the user specifies to "Keep one read for every 10 reads" (Figure 1), this means that for every 10 reads, the program will keep only 1 read. This is equivalent to keeping 10% of the data.
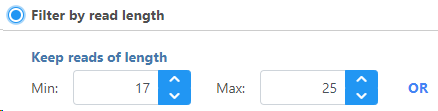
To Filter by read length, set the read length limits by choosing the minimum and maximum read length(s) to keep.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
44 | rates |
1 Comment
Melissa del Rosario
author: cignacio