The Feature distribution plot visualizes the distribution of features in a counts data node.
Running Feature distribution
To run Feature distribution:
- Click a counts data node
- Click the QA/QC section of the toolbox
- Click Feature distribution
A new task node is generated with the Feature distribution report.
Feature distribution plot configuration
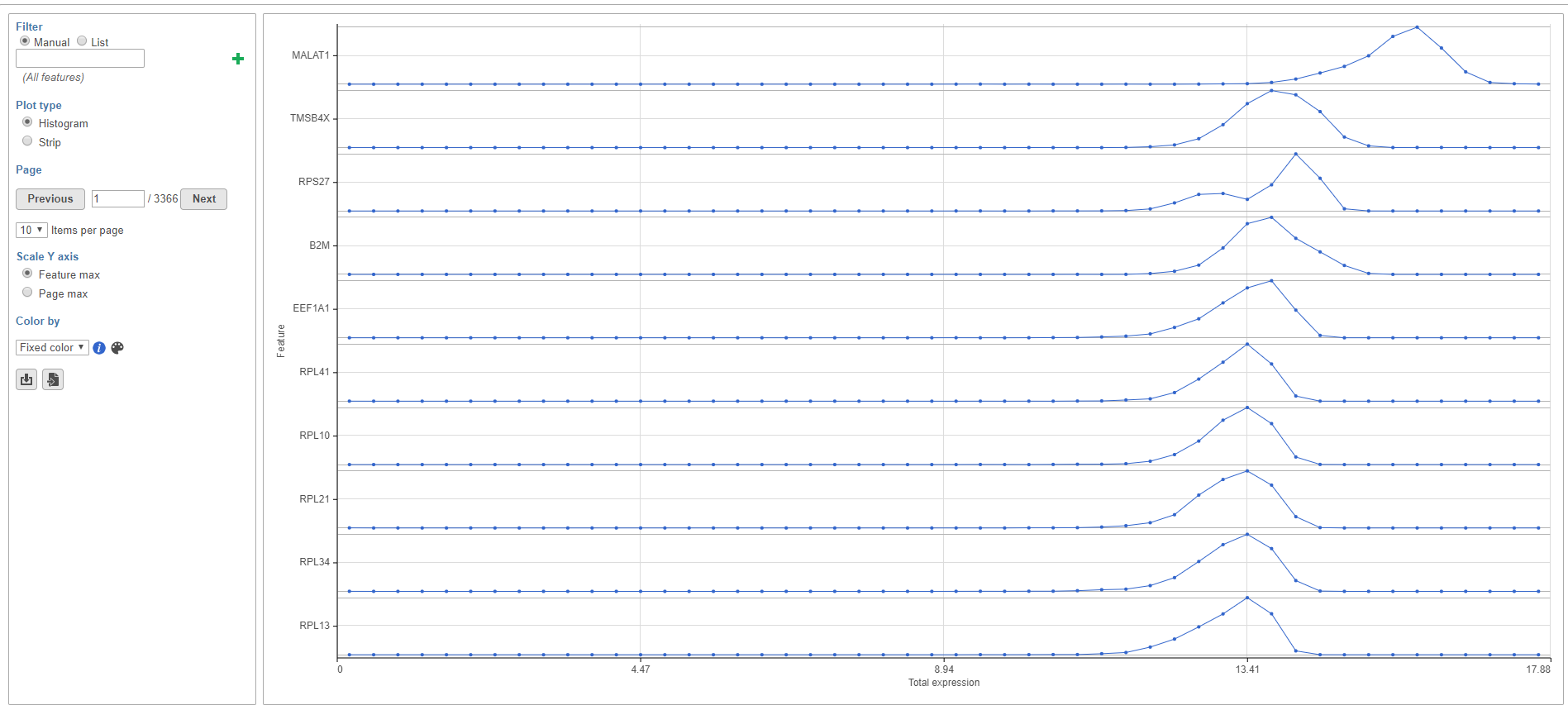
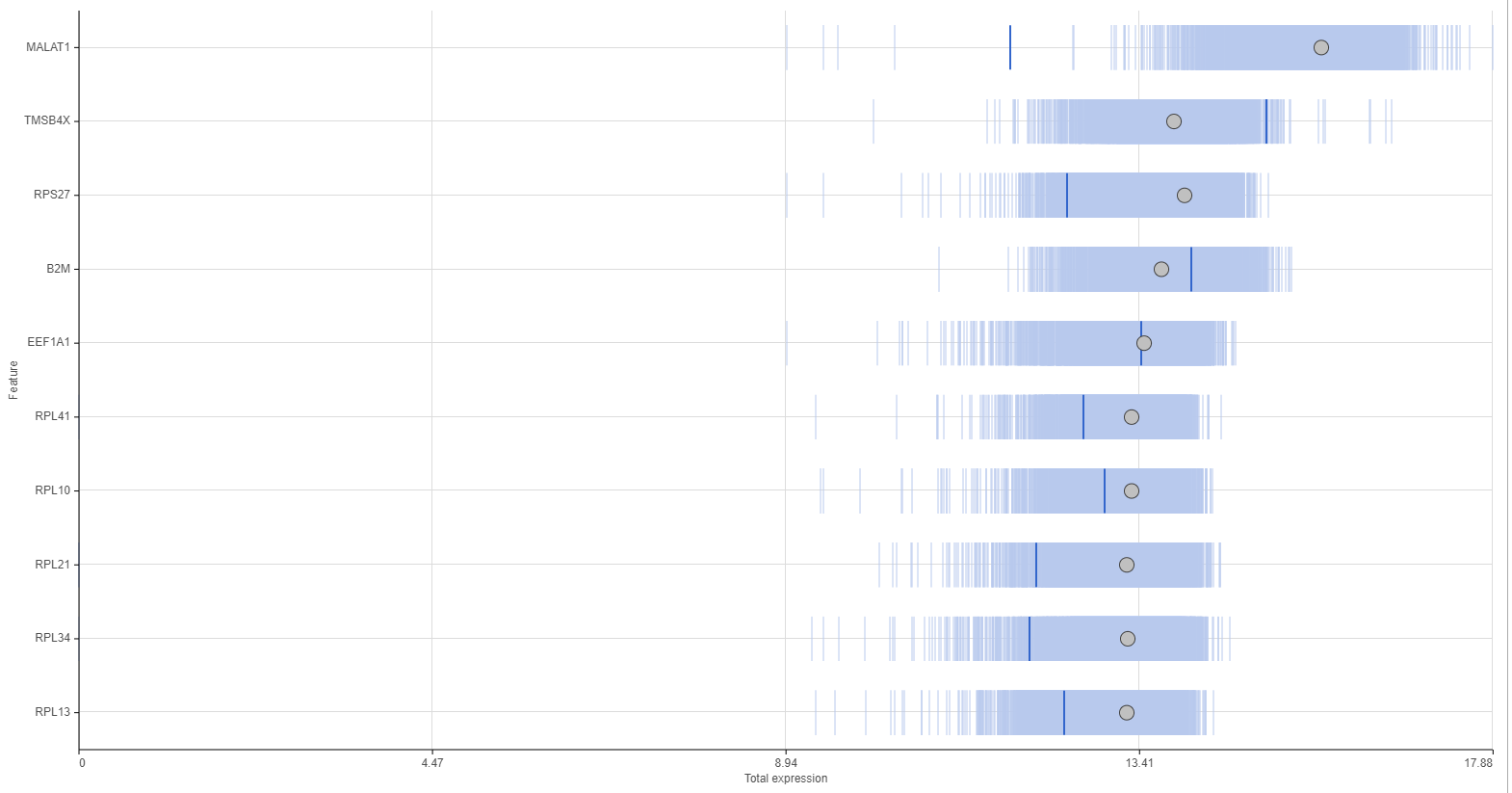
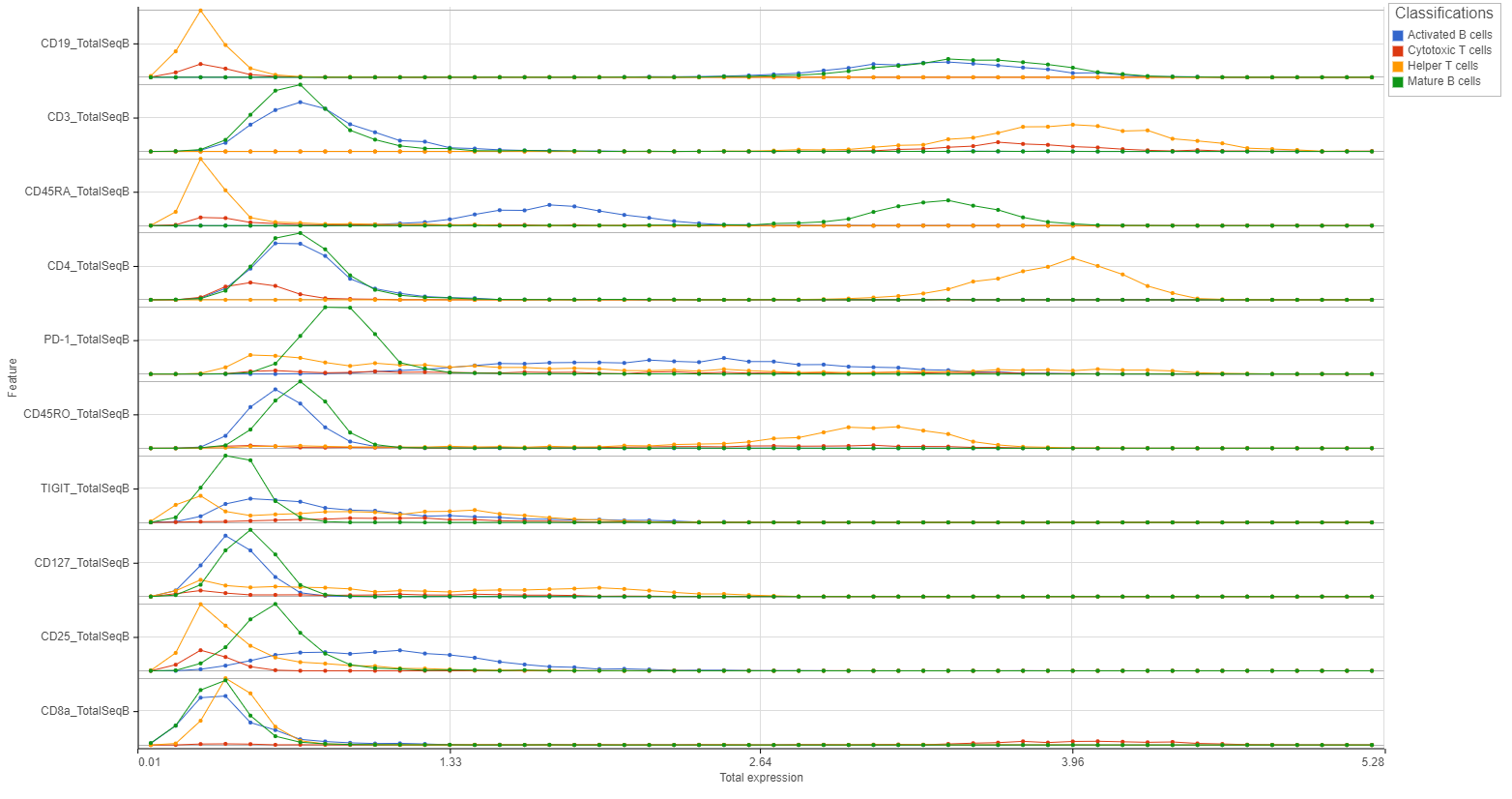
The Feature distribution task report plots the distribution of all features (genes or transcripts) in the input data node with one feature per row (Figure 1). Features are ordered by average value in descending order.
The plot can be configured using the panel of the left-hand side of the page.
Filter
Using the filter, you can choose which features are shown in the task report.
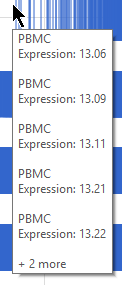
The Manual filter lets you type a feature ID (such as a gene symbol) and filter to matching features by clicking . You can add multiple feature IDs to filter to multiple features (Figure 2).
The List filter lets you filter to the features included in a feature list. To learn more about feature lists in Partek Flow, please see List management.
Plot type
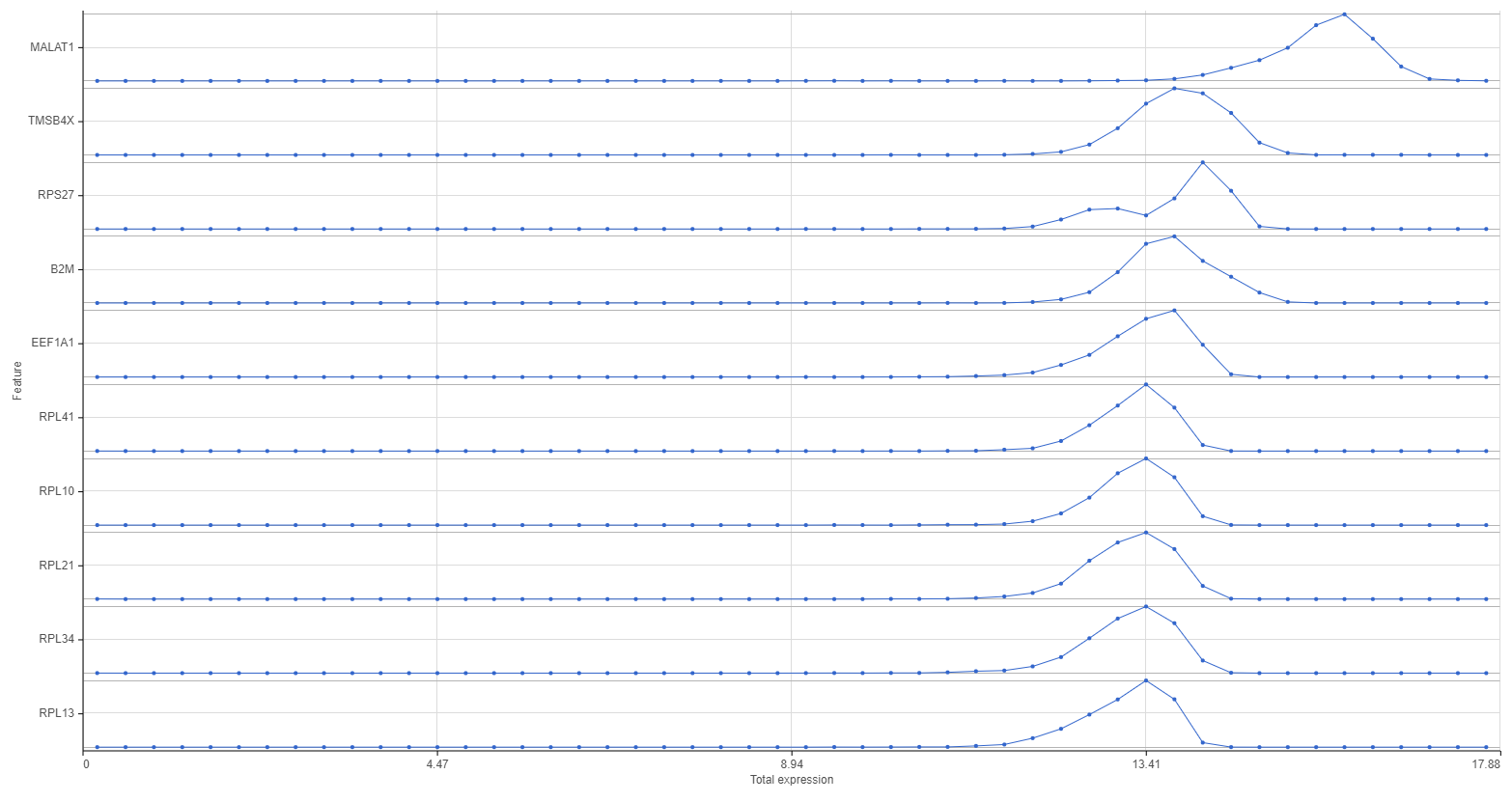
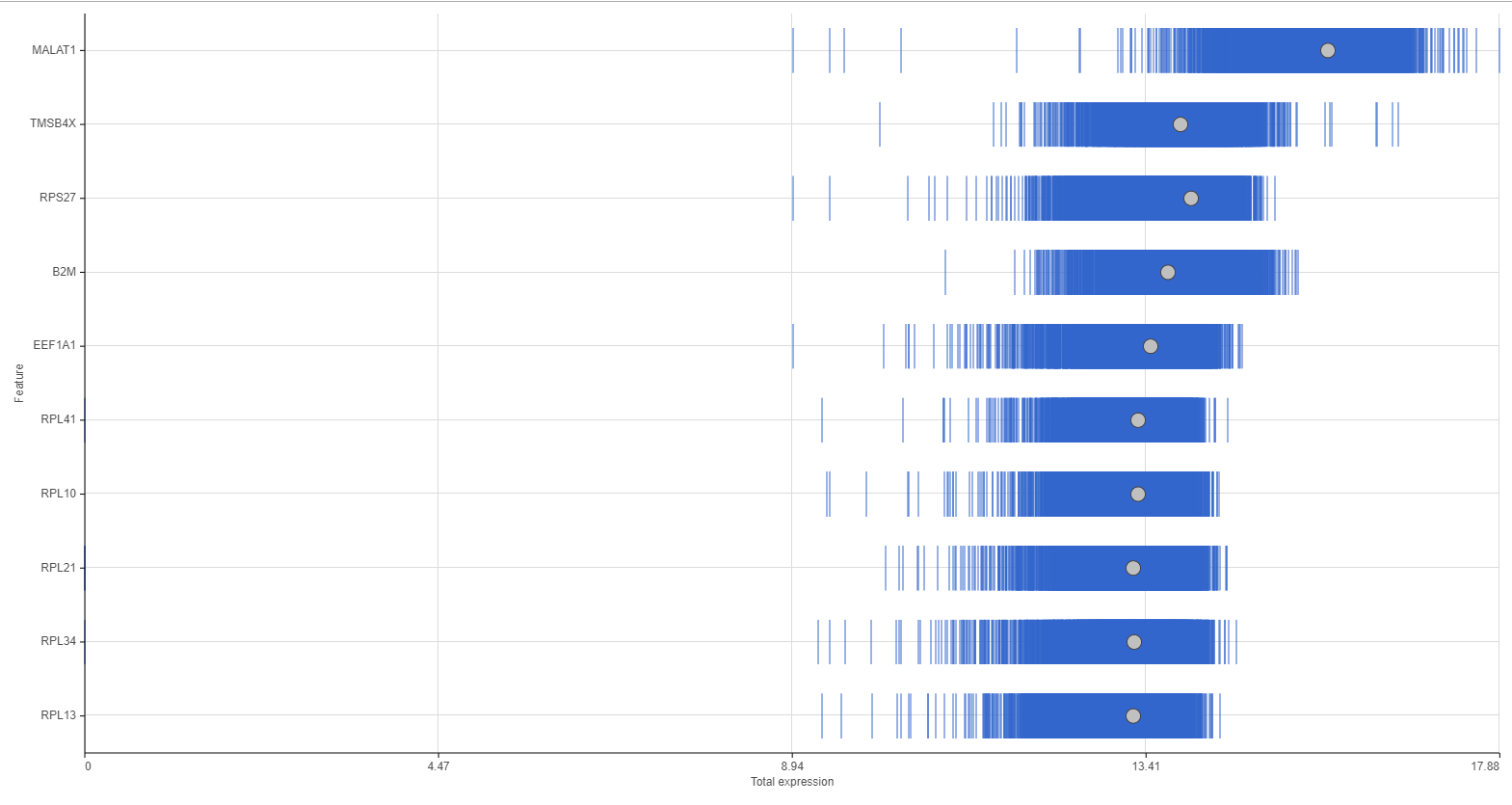
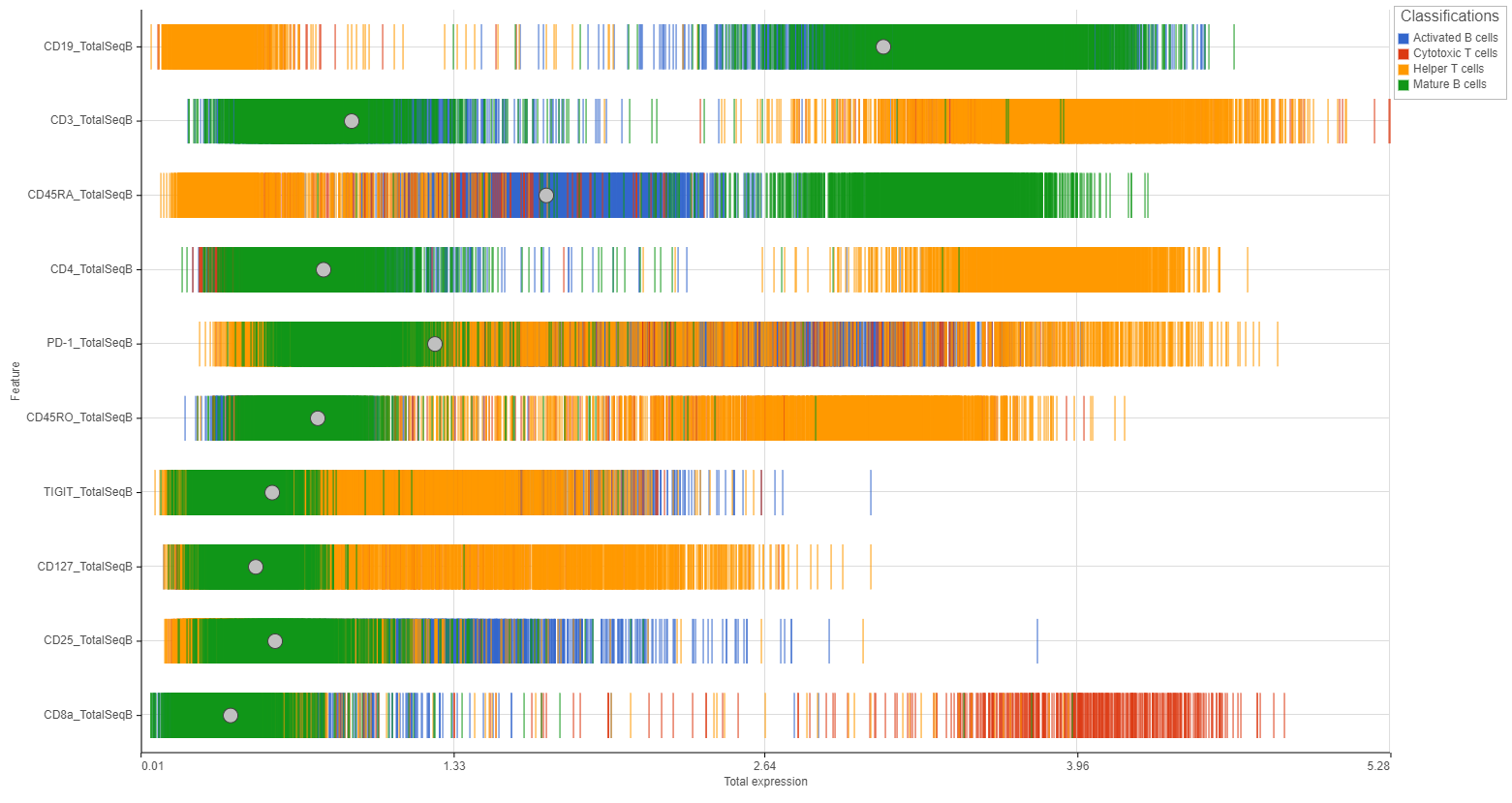
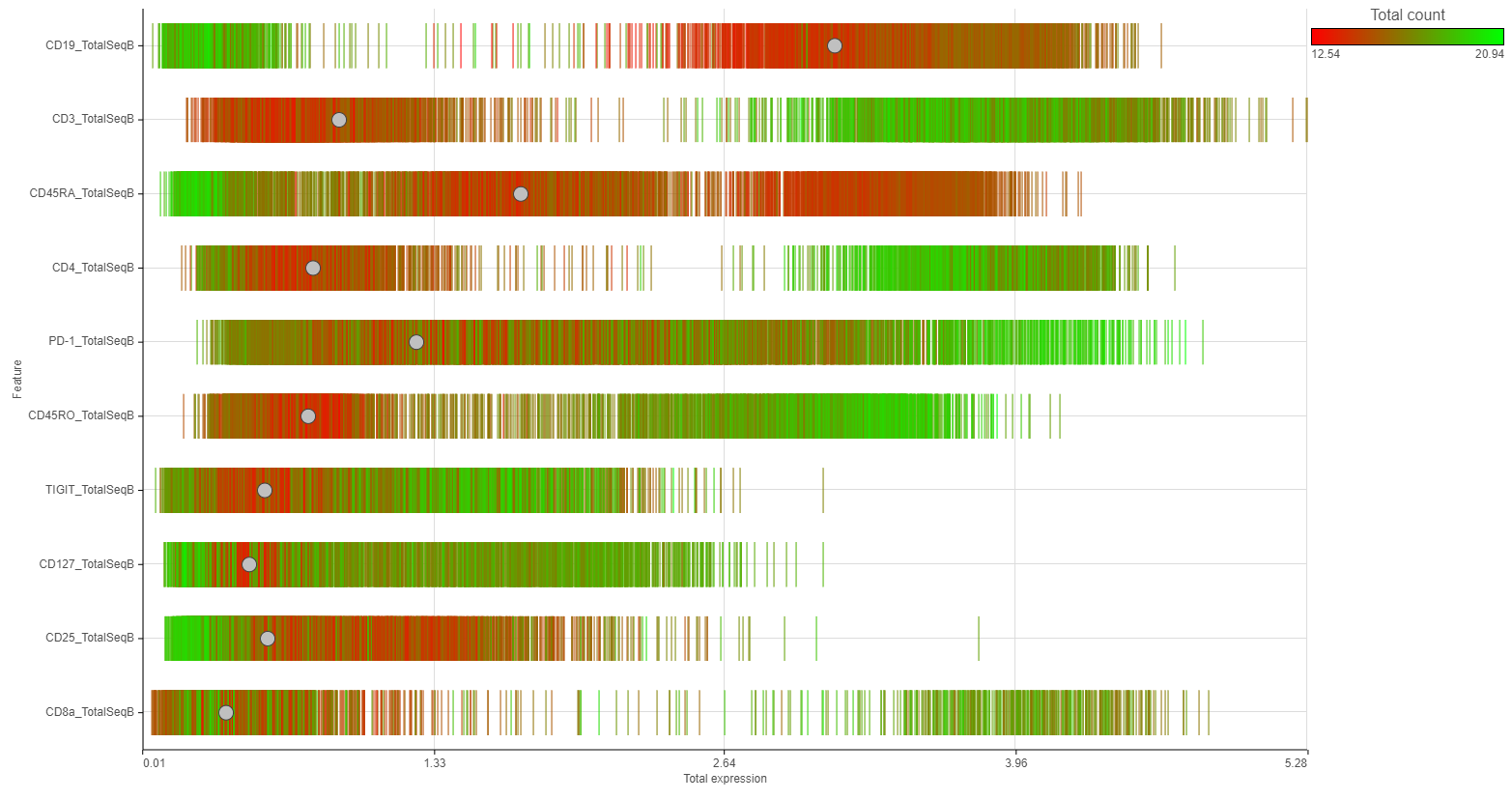
Distributions can be plotted as histograms, with the x-axis being the expression value and the y-axis the frequency, or as a strip plot, where the x-axis is the expression value and the position of each cell/sample is shown as a thin vertical line, or strip, on the plot (Figure 3).
To switch between plot types, use the Plot type radio buttons.
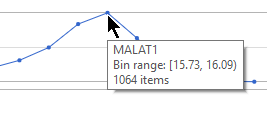
Mousing over a dot in the histogram plot gives the range of feature values that are being binned to generate the dot and the number of cells/samples for that bin in a pop-up (Figure 4).
Page
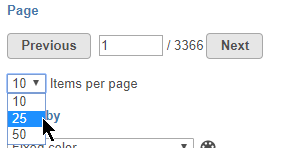
To navigate between pages, use the Previous and Next buttons or type the page number in the text field and click Enter on your keyboard.
The number of features that appear in the plot on each page is set by the Items per page drop-down menu (Figure 7). You can choose to show 10, 25, or 50 features per page.
Scale Y-axis
When Plot type is set to Histogram, you can choose to configure the Y-axis scale using the Scale Y-axis radio buttons. Feature max sets each feature plot y-axis individually. Page max sets the same y-axis range for every feature plot on the page, with the range determined by the feature with the highest frequency value.
Color by
You can add attribute information to the plots using the Color by drop-down menu.
For histogram plots, the histograms will be split and colored by the levels of the selected attribute (Figure 8). You can choose any categorical attribute.


| Your Rating: |
    
|
Results: |
    
|
23 | rates |