If you have attribute information about your cells, you can use the Annotate cells task in Partek Flow to apply this information to the data. Once applied, these can be used like any other attribute in Partek Flow, and thus can be used for cell selection, classification and differential analysis.
To run Annotate cells:
- Click a Single cell counts data node
- Click the Annotation/Metadata section in the toolbox
- Click Annotate cells
You will be prompted to specify annotation input options:
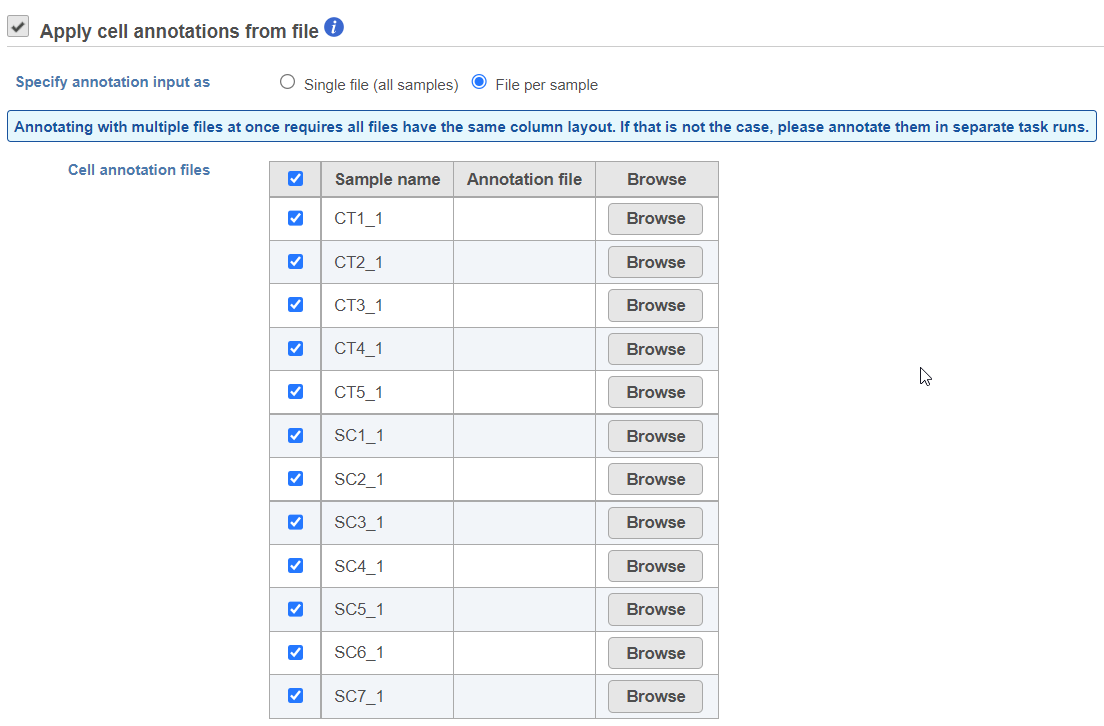
- Single file (all sample): it requires one .txt file for all your cells in all samples, each row in the file represents a barcode, at least one barcode column which will match the barcodes in your data. It also requires an column contains sample ID which will match the sample name in the data tab of your project.
- File per sample: it requires the format of all of the annotation files to be the same. Each file has barcodes on rows, it requires one barcode column that will match the barcodes in your data in that sample. All files have to have the same set of column, column headers are case sensitive.
You can pick the file for each sample from the Partek Flow server, you have to specify annotation files for all the samples in the dailog (Figure 1).
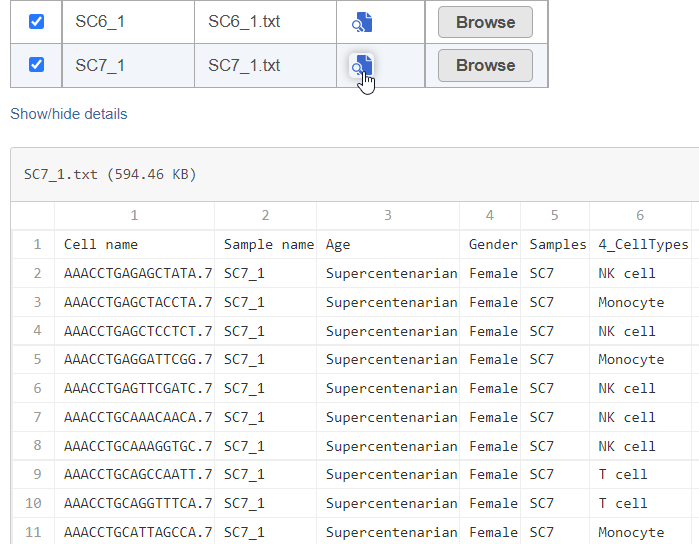
To view a preview of the files, click Show Preview (Figure 2).
- Click Next to continue
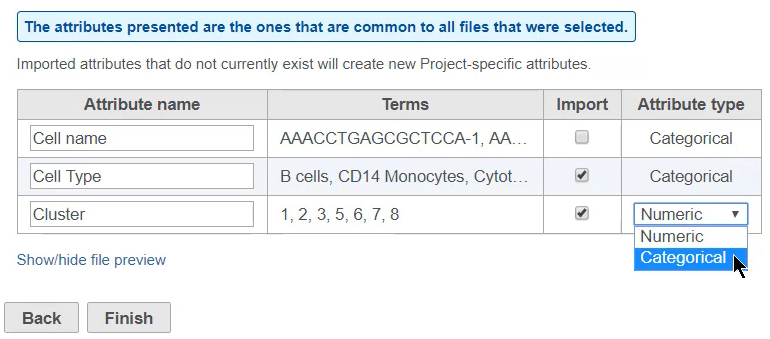
The next dialog page previews the attributes found in the annotations text file (Figure 3).
You can choose which attributes to import using the check-boxes, change the names of attributes using the text fields, and indicate whether an attribute with numbers is categorical or numeric.
- Click Finish to import the attributes.
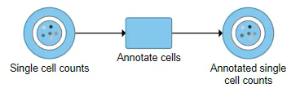
A new data node, Annotated single cell counts, will be generated (Figure 4).
You annotations will be available in downstream analysis tasks.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |