The Trim bases task is used to trim bases from the 5'-end or 3'-end of the reads. The most obvious reason for Trim bases is to trim away poor quality bases from the read prior to alignment because these can potentially affect alignment rate.
The task allows user to trim reads in different ways (Figure 1), including:
- Trim bases based on quality score
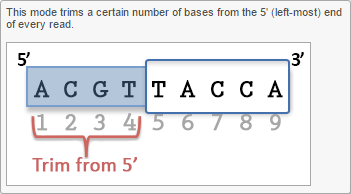
- Trim bases from 3'-end
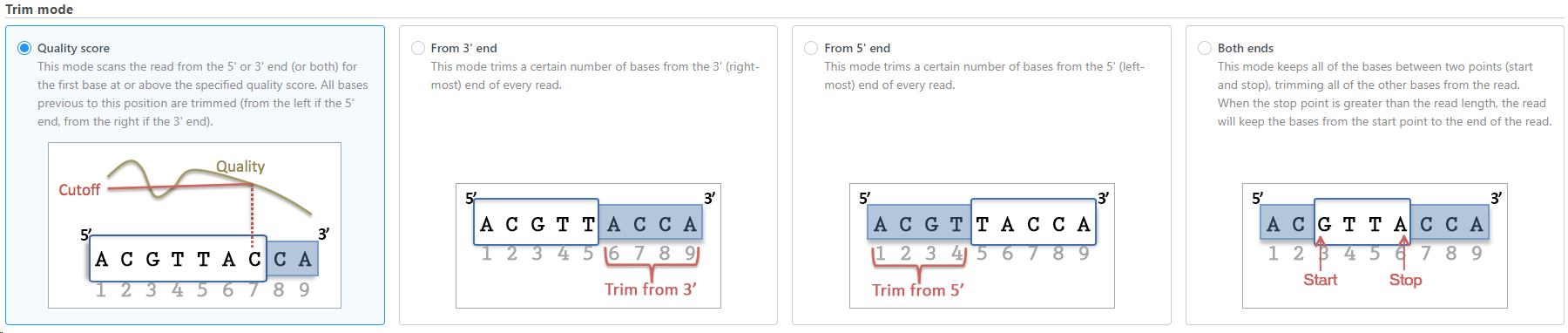
- Trim bases from 5'-end
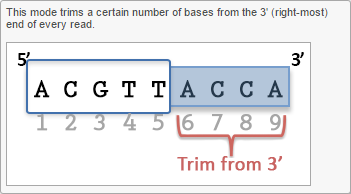
- Trim bases from both ends
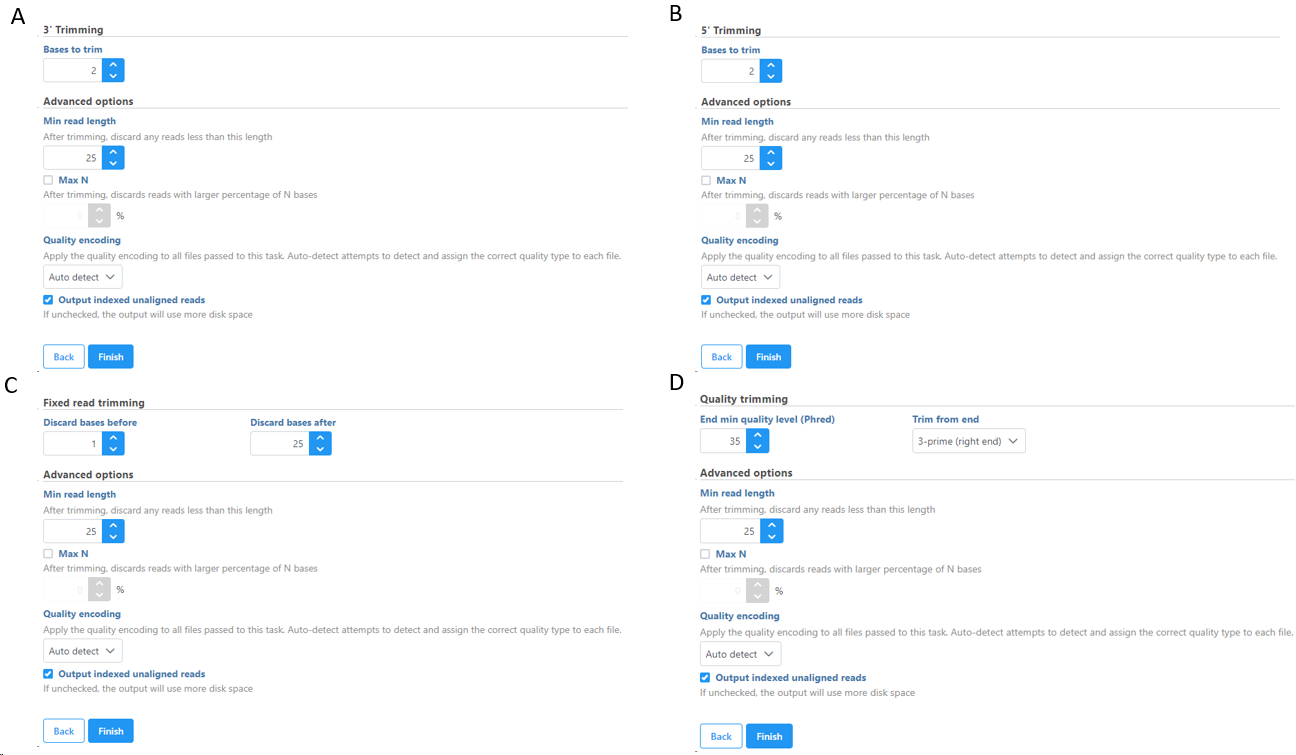
Trim bases from 5'-or 3'-end (Figures 2-3) allows a fixed number of bases to be trimmed away from the 5'- or 3'-end of the reads. These two functions are useful for when your read length is constant. This is not recommended if the read length is not constant, since good quality bases from shorter reads are likely trimmed away by these functions.
Trim bases from both ends (Figure 4) allows user to keep only bases from a fixed start and end position of the reads. This is particularly useful if poor quality bases are observed on both ends of the read. So instead of performing trim bases successively from the 5'- and 3'-end, the trim bases will only be performed once by trimming from both ends.
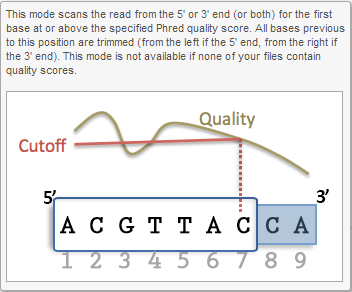
Trim bases based on quality score (Figure 5) is probably the most useful function to trim poor quality bases from the 5'- or 3'-ends of reads. This function allows dynamic trimming of bases depending on quality score. The trimming can be done from either 5'-end, 3'-end or both ends of the reads. The function evaluates each base from the end of the read and trims it away until the last base has a quality score greater than the specified threshold. For an extensive evaluation of read trimming effects on Illumina NGS data analysis, see Del Fabbro et. al. [1].
Advanced options
In some cases, the reads that result from base trimming can have very short read lengths and thus are not recommended for alignment. Thus, Partek® Flow® Flow provides the option to set a Min read length after base trimming. This discards reads that are shorter than the set length.
Also, reads could have a high percentage of N's or ambiguous bases. Thus, the Max N setting is available to discard reads with %Ns higher than the set threshold
The Quality encoding option refers to the Phred quality score encoded within the FASTQ input file. The list of available options are: Phred+33, Phred+64, Solexa+64 and Integers. Selecting Auto-detect will determine whether the quality encoding is Phred+33 or Phred+64. For Solexa data, you will need to select Solexa+64. For most of datasets, auto-detect option works very well with a few exception cases where the base quality score falls into the grey zone (ambiguous zone) of Phred+33 and Phred+64 score. However, if the quality-encoding scheme is known, we recommend to selecting the encoding format directly from the quality encoding list.
Figure 6 shows the options available for all the different selection of Trim bases function. Note the default Min read length is 25bp. For micro RNA sequencing data, this default Min read length needs to be set to a smaller value (we recommend 15) to account for mature microRNAs.
Trim Bases Task Details Page
The Task Details page for Trim bases can be accessed by selecting the task node Trim bases, and subsequently selecting Task Details from the Task results section. In the Task details page, several sections are available:
- General task information: contains information such as the task name, owner, status, submitted time, start, end and duration of the task
- Output Files: contains the description of each output file. If you roll-over your mouse cursor to the file name, you will get the exact location of the file on the server. If you click on the file name, you will have the option to view up to 999 lines of the raw data. You can also download the file from the server.
- Input Files: contains the information of input files. This section lists down all the input files used in the Trim bases task.
- Input Parameters: contains the parameters used for running Trim bases function. This section tells what option has been selected for the Trim bases task. It includes all the parameters used for the task, such as minimum read length, maximum percentage of N's base, quality encoding, quality score threshold (if applicable) and how trimming is performed.
- Command Lines: shows the commands used for running Trim bases function by the software Partek Flow
Trim Bases Task Report Page
The Trim bases Task Report page can be accessed by selecting either the Trim bases task node or Trimmed reads data node and then selecting the Task Report from the Task results section of the context sensitive menu. There is a link at the bottom of the page to directly go to the Task Details page. The page displays the following components:
- Summary table: gives the total number of reads in each sample, the total number of reads trimmed (i.e. with at least one base trimmed from the read), total number of reads removed (due to Min read length and Max N parameters), the average number of bases trimmed per read, the average read quality before trim bases and finally the average read quality after trim bases.
- Stacked bar-chart: shows percentage of untrimmed reads, trimmed reads and removed reads are shown in a stacked bar-chart to compare all the samples.
- Average base quality score per position of trimmed reads: shows the average base quality score at each position of the trimmed reads for all samples in the project.
Trim Bases Output Files
The Trim bases function produces trimmed unaligned reads which is named as Trimmed Reads data node. The Trimmed Reads node will have the "trimmed" word appended to the filename. The Trimmed Reads data can be downloaded by selecting the Trimmed Reads node and then select Download data from the context sensitive menu. However, if you have access to the Partek Flow server, you can go to the Task Details page and identify the location of the output files from the Output Files section as described on the Trim Bases Task Details section above. The Trimmed Reads data node will have the same format as the raw data.
References
- Del Fabbro C, Scalabrin S, Moragante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS ONE. 2013; 8(12): e85024.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
43 | rates |
1 Comment
Melissa del Rosario
author: cignacio