This tutorial uses a spreadsheet generated after data import, but we will illustrate the steps used to import the data in this section.
- Select Copy Number from the Workflows drop-down menu
- Select Import Samples from the Copy Number workflow
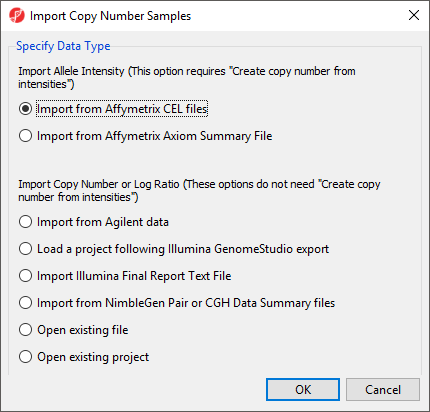
The import dialog will open (Figure 1).
For Affymetrix arrays, Partek Genomics Suite can import CEL files with allele intensity values and calculate copy number estimates from these intensities. For Agilent, Illumina, NimbleGen, or Affymetrix .CHP files, Partek Genomics Suite can import files containing calculated copy numbers or log ratios.
For this tutorial, we will not be importing CEL files.
- Select Cancel to close the import dialog
Later sections of this tutorial will address starting with copy number or log ratios and performing GC wave correction on Affymetrix CEL files.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
0 | rates |