Cell Ranger is a set of analysis pipelines that process Chromium single-cell RNA-seq output to align reads, generate feature-barcode matrices and perform clustering and gene expression analysis for 10X Genomics Chromium Technology[1].
Cell Ranger v6.0.0[2] has been wrapped In Partek® Flow® as Cell ranger task. It does not comprehensively cover all of the options and analysis cases Cell Ranger can handle for now, but converts FASTQ files from cellranger mkfastq and performs alignment, filtering, barcode counting, UMI counting. The output gene expression count matrix in .h5 format (both raw and filtered) then becomes the starting point for downstream analysis for scRNA-seq in Flow.
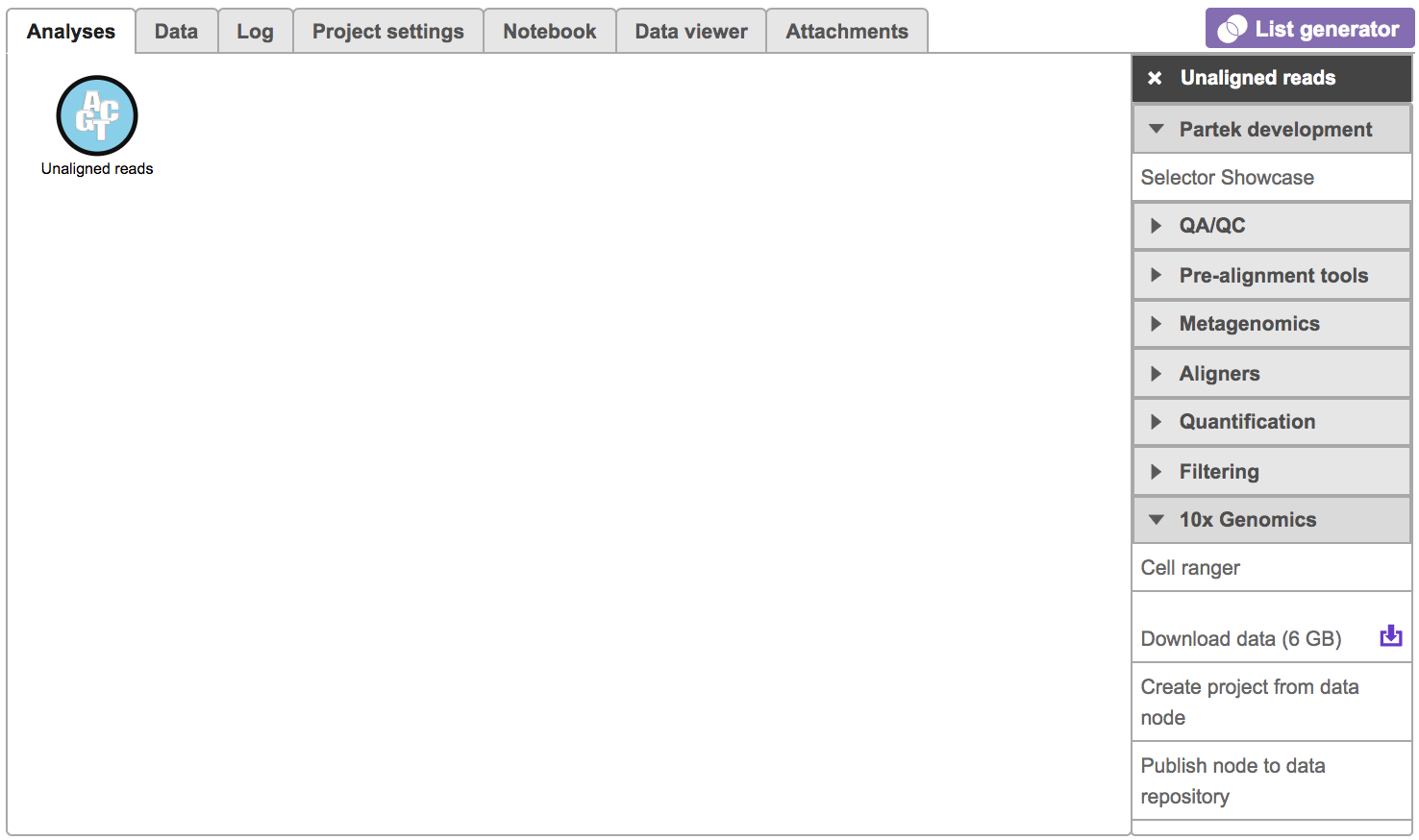
To run the ‘Cell ranger’ task in Flow, select ‘Unaligned reads’ datanode, then select ‘Cell ranger’ in the ‘10x Genomics’ section (Figure 1).
We recommend filtering to a set of genes you want to test for enrichment, but Gene set enrichment will run on any Feature list data node.
The background gene list is used as the list of possible genes. By default, this is the genes included in the selected gene set database. If your assay limits the genes that could be detected, you may want to specify a background list.
The gene sets available for the current Assembly are listed under the Gene set drop-down list. The assembly is automatically selected, if possible. If the assembly cannot be detected, you can specify it using a drop-down menu.
|
By default, the groups are defined by Gene Ontology (GO), a bioinformatics initiative to unify the representation of gene and gene product attributes across various species [1, 2].
Alternatively, selecting the Add gene ontology source from the Gene set drop down list option opens another dialog (Figure 3), where you can either Download gene set from Partek® (Recent GO database gene sets for human, mouse and rat are available) or Import gene set. The latter option takes you to the file browser, where you can point to the file that you want to use (not shown). Partek® Flow® accepts .gmt files as gene set inputs.
|
The result is stored under an Enrichment task node. To open it, double click on the node or select the respective Task report from the context sensitive menu.
Figure 4 shows an example Gene set enrichment task report. The table contains one gene set per row (Gene set column; the column entries are hyperlinks when using the distributed GO gene sets), with the category name in the Description column. The categories are ranked by the Enrichment score, which is the negative natural logarithm of the enrichment p-value (P-value column) derived from Fisher's exact test on the underlying contingency table. The higher the enrichment score, the more overrepresented the GO category is within the input list of significant genes. The columns can be searched by typing in the search term in the respective box (and hitting Enter), or sorted by selecting the double arrow icon ( ![]() ).
).
The contingency table (Figure 5) can be displayed by selecting the View gene breakdown chart icon on the right (![]() ). The term "list" refers to the list of significant genes, while the term "set" refers to the respective GO category. The first row of the contingency table is also seen in the report, namely the Genes in list and Genes not in list columns.
). The term "list" refers to the list of significant genes, while the term "set" refers to the respective GO category. The first row of the contingency table is also seen in the report, namely the Genes in list and Genes not in list columns.
|
The View extra details (![]() ) button provides additional information on the GO category (Figure 6). In addition to the details already given in the report, a full list of Genes in list and Genes not in list can be inspected and downloaded (Download data) to the local computer as a text file.
) button provides additional information on the GO category (Figure 6). In addition to the details already given in the report, a full list of Genes in list and Genes not in list can be inspected and downloaded (Download data) to the local computer as a text file.
|
As previously mentioned, if you are using the GO gene sets distributed by Partek, the GO identifiers in the first column are hyperlinks to the Gene Ontology web-site entries (an example shown in Figure 7).
|
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/6.0/release-notes