Partek® Flow® can import a wide variety of data types including raw counts, matrices, microarray, variant call files as well as unaligned and aligned NGS data.
The following file types are valid and will be recognized by the Partek Flow file browser.
|
|
|
In cases where paired end fastq data is present, files will also be automatically recognized and their paired relationship will be maintained throughout the analysis.
Matching on paired end files is based on file names: every character in both file names must match, except for the section that determines whether a file is the first or the second file. For instance, if the first file contains "_R1", "_1", "_F3", "_F5" in the file name, the second file must contain something in the lines with the following: "_R2", "_2", "_F5", "_F5-P2", "_F5-BC", "_R3", "_R5" etc. The identifying section must be separated from the rest of the filename with underscores or dots. If two conflicting identifiers are present then the file is treated as single end. For example, s_1_1 matches s_1_2, as described above. However, s_2_1 does not mate with s_1_2 and the files will be treated as two single-end files.
Apart from paired-end data, files with conventional filename suffixes that indicate that they belong to the same sample are consolidated. These suffixes include:
Dates
in the form "####-##-##" preceded or followed by a period or underscore
Set number
of the form "_###" from the end
The file browser is used to transfer files to the server so that these files can be added to a project for analysis. If you are importing a Bioproject from GEO/ENA or using URLs for data import, there is no need to transfer the files to the server.
To access the file browser and upload data to the server, use any of these options:
Using the file browser to transfer files to the server:
 to access the file browser
to access the file browser to add files from your machine
to add files from your machine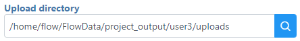 should be specified, known, and distinguishable for project file management. You will return to this directory and access the files to import them into a project
should be specified, known, and distinguishable for project file management. You will return to this directory and access the files to import them into a project to complete the file upload
to complete the file uploadFile size displayed in the table is binary format, not decimal format (e.g. GB displayed in the table is gigibyte not gigabyte. 1 gigibyte is 1,073,741,824 bytes. 1 gigabyte is 1,000,000,000 bytes. 1 gigibyte is 1.074 gigabytes).
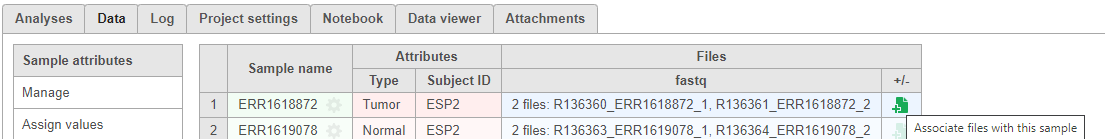
There are projects with more than one file type, such as single cell multi-omic assays that generate protein and RNA together. In these cases, files need to be associated with each other if starting in fastq format. If we start with processed data, there is no need to associate these files.
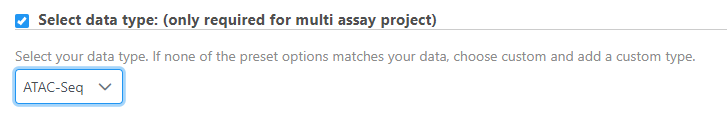
If this step is skipped, the data type can be changed after import by right clicking the data node.