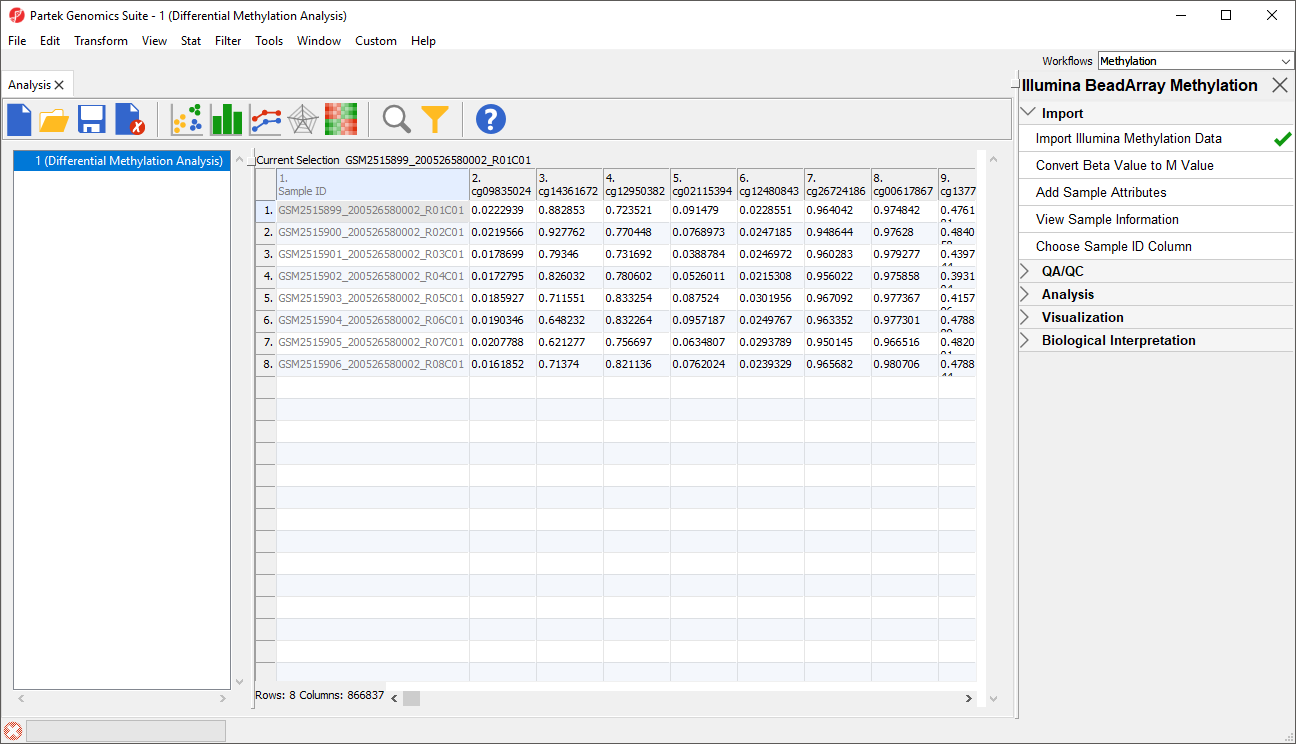
Each row of the spreadsheet (Figure 1) corresponds to a single sample. The first column is the names of the .idat files and the remaining columns are the array probes. The table values are β-values, which correspond to the percentage methylation at each site. A β-value is calculated as the ratio of methylated probe intensity over the overall intensity at each site (the overall intensity is the sum of methylated and unmethylated probe intensities).
|
An alternative metric for measurement of methlyation levels are M-values. β-values can be easily converted to M-values using the following equation:
M-value = log2( β / (1 - β))
An M-value close to 0 for a CpG site indicates a similar intensity between the methylated and unmethylated probes, which means the CpG site is about half-methylated. Positive M-values mean that more molecules are methylated than unmethylated, while negative M-values mean that more molecules are unmethylated than methylated. As discussed by Du and colleagues, the β-value has a more intuitive biological interpretation, but the M-value is more statistically valid for the differential analysis of methylation levels.
Because we are performing differential methylation analysis, we need to convert our data to from β-values to M-values.
The original data (β-values) will be overwritten.
Before we can perform any analysis, the study samples need to be organized into their experimental groups.
|
The Create categorical attribute dialog allows us to create groups for a categorical attribute. By default, two groups are created, but additional groups can be added.
| Sample ID | Group Name |
|---|---|
| GSM2515899_200526580002_R01C01 | Primed+shCTRL |
| GSM2515900_200526580002_R02C01 | Primed+shCTRL |
| GSM2515901_200526580002_R03C01 | Naive+shCTRL |
| GSM2515902_200526580002_R04C01 | Naive+shCTRL |
| GSM2515903_200526580002_R05C01 | Naive+shPOU5F1 |
| GSM2515904_200526580002_R06C01 | Naive+shPOU5F1 |
| GSM2515905_200526580002_R07C01 | Naive+shNANOG |
| GSM2515906_200526580002_R08C01 | Naive+shNANOG |
There should now be four groups with two samples in each group (Figure 4).
|
A new column s been added to spreadsheet 1 (Differential Methylation Analysis) with the state and shRNA treatment of each sample (Figure 6).
|
|