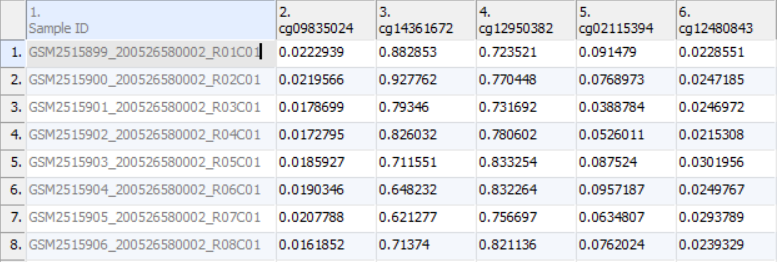
Once the .idat files are imported, you will be facing the Scatter Plot. To move forward with the tutorial, switch to the Analysis tab and take a look at the top level (i.e. initial) spreadsheet (Figure 1). Samples identifiers are the names of the .idat files as shown by the Sample ID column while all the remaining columns are the array probes.
|
As discussed in the previous chapter, the values in the cells are normalized β-values, which correspond to the percentage of methylation at each site and are calculated as the ratio of methylated probe intensity over the overall intensity at each site (the overall intensity is the sum of methylated and unmethylated probe intensities). An alternative metric for measurement of methlyation levels are M-values. β-values can be easily converted to M-values using the following equation:
M-value = log2( β / (1 - β))
Here's the interpretation: a M-value close to 0 indicates a similar intensity between the methylated and unmethylated probes, which means the CpG site is about half-methylated. Positive M-values mean that more molecules are methylated than unmethylated, while negative M-values mean that more molecules are unmethylated than methylated. As discussed by Du and colleagues, the β-value has a more intuitive biological interpretation, but the M-value is more statistically valid for the differential analysis of methylation levels.
Therefore, the differential methylation analysis of the tutorial data will be performed on the M-values; select Convert Beta Value to M Value from the workflow. Note that the original data (β-values) will be overwritten.
Before proceeding to exploratory data analysis, the study samples need to be organized in four groups, two biological replicates each. In the Import section of the workflow, select Add Sample Attributes and, in the next dialog (Figure 2), Add a Categorical Attribute (and then OK).
|
The following dialog is Create Categorical Attribute (Figure 3). The Attribute name filed specifies the name of the new column (attribute), while the groups (levels of the attribute) are specified in the Group Name fields. By default, two groups are created, but additional ones can be added using the New Group button. Finally, drag and drop the samples from the Unassigned list to their matching groups.
|
For this analysis, specify Attribute name as HPSC and then create the groups and add samples to them using the table below.
| Sample ID | Group Name |
|---|---|
GSM2515899_200526580002_R01C01 | Primed |
GSM2515900_200526580002_R02C01 | Primed |
GSM2515901_200526580002_R03C01 | Naive |
GSM2515902_200526580002_R04C01 | Naive |
GSM2515903_200526580002_R05C01 | shPOU5F1 |
GSM2515904_200526580002_R06C01 | shPOU5F1 |
GSM2515905_200526580002_R07C01 | shNANOG |
GSM2515906_200526580002_R08C01 | shNANOG |
Push OK. When asked to add another categorical attribute, say No, and the confirm that you would like to save the spreadsheet (Yes).
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
|