GO ANOVA output is very similar to standard ANOVA output except each row in the resulting sheet contains statistical results from a single GO functional group rather than a single gene. Columns can be broken down into four sections:
- Annotations contain detail about the category being considered
- ANOVA results contain the significance of the effect of the factors in the model
- Contrast results contain significance and fold change of the difference between groups compared via contrast
- F-ratios display the significance of the factors in the ANOVA model
Annotations
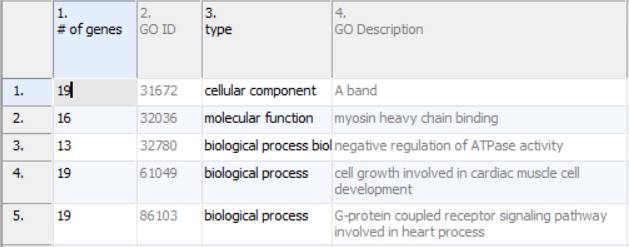
Annotations will take up the first four columns of the results sheet (Figure 1). The first column (# of genes) is the number of genes in the GO category. Specifically, this is not necessarily the number of unique genes in the category; depending on the technology, it can be the number of probes or probe sets on the microarray whose targets fall into the GO category. Genes targeted more than once will be counted more than once. The second column (GO ID) is the unique numeric identifier of the GO category; it is sometimes useful for searching with when the GO category has a very long name. The third column is the type of the GO category, while the fourth column (GO Description) is the name of the GO category.
When right click on any row header to choose Create Gene List , a new spreadsheet will be generated, it contains a list of genes (probes/probesets) within the selected GO category.
ANOVA Results
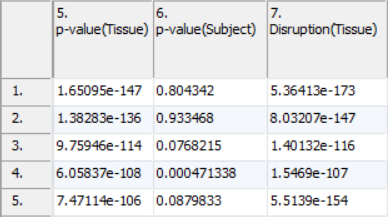
ANOVA results will include a column for each factor in the setup (Figure 2). A column with the name of the factor or interaction followed by p-value will contain how significant the effect of the variable is on the data. A lower p-value corresponds with a more significant effect. For example, a p-value of 0.1 for tissue means that given the difference between the tissue and the inherent variability of the measurements of the genes in the functional group, there is a 10% likelihood that the tissues are equivalent. A p-value of 0 occurs when the value is too small to be displayed. This can be caused by a very low estimate of inherent variability due to either a very small number of replicates or severely unbalanced data.
In the example experiment, a low p-value for tissue would imply the functional group is differentially expressed across tissues.
A low p-value for an interaction implies that the effect of one factor on the other is significant. In the example dataset, no interactions between two main variables were included as factors. To illustrate what the interaction p-value would mean, consider the case that a drug compound and a control injection were dosed over several time points and an interaction between injection compound and time point was included in the GO ANOVA. A low p-value for the drug-time point interaction corresponds to the effect of drug on the functional group being altered with time.
A column will also be present for each factor placed in the Disruption Factor(s) box. This column will have the header Disruption(Factor name). A low p-value in this column corresponds to the different states presenting with different gene patterns within the functional group. For functional groups containing only a single gene, no value will be present as the pattern cannot change. In the example experiment, a low p-value for the Disruption(Tissue) represents function categories which have different genes operating in the heart and in the brain.
Contrast Results
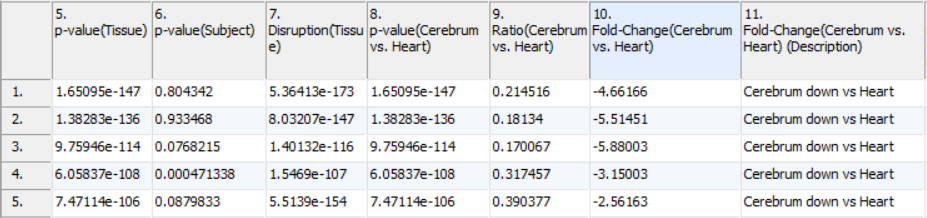
Contrast results include four columns for each of the comparisons declared during GO ANOVA setup. The first column contains the p-value representing the significance of the difference between the two categories. The second column contains the ratio between the two groups where increases are represented as greater than one and decreases are represented as values between zero and one. The third column is the fold change of the functional group between the two categories where increases are greater than one and decreases are less than negative one. The fourth column contains a plain text description of the direction of the fold change. Fold changes and ratios represent the average change in the functional category. In the example, a contrast was run comparing expression in the cerebral tissue to the heart tissue (Figure 3). As these were the only tissues, the p-values are identical to those in column 5. While the p-value column shows which groups are differentially expressed between the tissues, the fold change columns allow us to see by how much they are differentially expressed. Using the sign of the fold change, or the description column, you can see which categories are increased in brain and which are increased in heart.
F-Ratios
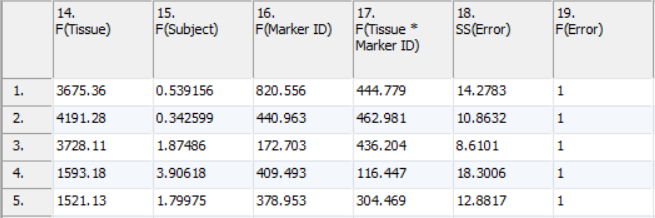
F-ratios (Figure 4) are used in the computation of p-values. The values in the columns can safely be ignored by most users; there are exceptional cases when the F-ratios may be informative. To see the general significance of the factors included in the model, a Sources of Variation plot can be computed from these values from the View menu (or the Workflow). The higher the average F-ratio, the more important the factor is to the model on average.
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.


| Your Rating: |
    
|
Results: |
    
|
33 | rates |